[English] 日本語
 Yorodumi
Yorodumi- PDB-6pw2: Structural Basis for Cooperative Binding of EBNA1 to the Epstein-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6pw2 | ||||||
|---|---|---|---|---|---|---|---|
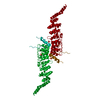
| Title | Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication | ||||||
 Components Components |
| ||||||
 Keywords Keywords | viral protein/dna / EBNA1 / DNA binding protein / Epstein-Barr Virus / viral protein / viral protein-dna complex | ||||||
| Function / homology |  Function and homology information Function and homology informationhost cell PML body / viral latency / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / symbiont-mediated disruption of host cell PML body / regulation of DNA replication / enzyme-substrate adaptor activity / endonuclease activity / symbiont-mediated suppression of host NF-kappaB cascade / DNA-binding transcription factor activity / positive regulation of DNA-templated transcription / DNA binding Similarity search - Function | ||||||
| Biological species |  Epstein-Barr virus (Epstein-Barr virus) Epstein-Barr virus (Epstein-Barr virus) Human herpesvirus 4 strain B95-8 (Epstein-Barr virus (strain B95-8)) Human herpesvirus 4 strain B95-8 (Epstein-Barr virus (strain B95-8)) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.01 Å MOLECULAR REPLACEMENT / Resolution: 3.01 Å | ||||||
 Authors Authors | Messick, T.E. / Malecka, K.A. / Lieberman, P.M. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: J.Virol. / Year: 2019 Journal: J.Virol. / Year: 2019Title: Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication. Authors: Malecka, K.A. / Dheekollu, J. / Deakyne, J.S. / Wiedmer, A. / Ramirez, U.D. / Lieberman, P.M. / Messick, T.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6pw2.cif.gz 6pw2.cif.gz | 683.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6pw2.ent.gz pdb6pw2.ent.gz | 558.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6pw2.json.gz 6pw2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6pw2_validation.pdf.gz 6pw2_validation.pdf.gz | 503.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6pw2_full_validation.pdf.gz 6pw2_full_validation.pdf.gz | 529.7 KB | Display | |
| Data in XML |  6pw2_validation.xml.gz 6pw2_validation.xml.gz | 48.3 KB | Display | |
| Data in CIF |  6pw2_validation.cif.gz 6pw2_validation.cif.gz | 67.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pw/6pw2 https://data.pdbj.org/pub/pdb/validation_reports/pw/6pw2 ftp://data.pdbj.org/pub/pdb/validation_reports/pw/6pw2 ftp://data.pdbj.org/pub/pdb/validation_reports/pw/6pw2 | HTTPS FTP |
-Related structure data
| Related structure data |  1b3tS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 16008.498 Da / Num. of mol.: 8 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Epstein-Barr virus (strain B95-8) (Epstein-Barr virus (strain B95-8)) Epstein-Barr virus (strain B95-8) (Epstein-Barr virus (strain B95-8))Strain: B95-8 / Gene: EBNA1, BKRF1 / Production host:  #2: DNA chain | Mass: 19128.277 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: CTAACCCTAATTCAATAGCATATGTTACCCAACGGGAAGCATATGCTATCGAATTAGGGTTA Source: (synth.)  Human herpesvirus 4 strain B95-8 (Epstein-Barr virus (strain B95-8)) Human herpesvirus 4 strain B95-8 (Epstein-Barr virus (strain B95-8))#3: DNA chain | Mass: 19084.285 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: CTAACCCTAATTCAATAGCATATGTTACCCAACGGGAAGCATATGCTATCGAATTAGGGTTA Source: (synth.)  Human herpesvirus 4 strain B95-8 (Epstein-Barr virus (strain B95-8)) Human herpesvirus 4 strain B95-8 (Epstein-Barr virus (strain B95-8))#4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.02 Å3/Da / Density % sol: 59.27 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 6.75 / Details: 200 mM sodium malonate, 24% PEG 3350 / PH range: +/- 0.25 / Temp details: +/- 2 degrees |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: Cryostream / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 1 Å / Beamline: 19-ID / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Nov 19, 2015 |
| Radiation | Monochromator: Si (111) Rosenbaum-Rock double-crystal monochromator Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3→50 Å / Num. obs: 29088 / % possible obs: 99.6 % / Redundancy: 10.6 % / Rsym value: 0.159 / Net I/σ(I): 8.1 |
| Reflection shell | Resolution: 3→3.05 Å / Redundancy: 8.5 % / Mean I/σ(I) obs: 0.16 / Num. unique obs: 2264 / Rsym value: 2.682 / % possible all: 98.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1B3T Resolution: 3.01→40.9 Å / Cross valid method: FREE R-VALUE Details: TLS record as of below: tls = "chain 'A' and (resid 461 through 477 )" tls = "chain 'A' and (resid 478 through 491 )" tls = "chain 'A' and (resid 492 through 504 )" tls = "chain 'A' and ...Details: TLS record as of below: tls = "chain 'A' and (resid 461 through 477 )" tls = "chain 'A' and (resid 478 through 491 )" tls = "chain 'A' and (resid 492 through 504 )" tls = "chain 'A' and (resid 505 through 547 )" tls = "chain 'A' and (resid 548 through 560 )" tls = "chain 'A' and (resid 561 through 583 )" tls = "chain 'A' and (resid 584 through 599 )" tls = "chain 'A' and (resid 600 through 607 )" tls = "chain 'B' and (resid 461 through 477 )" tls = "chain 'B' and (resid 478 through 489 )" tls = "chain 'B' and (resid 490 through 513 )" tls = "chain 'B' and (resid 514 through 547 )" tls = "chain 'B' and (resid 548 through 560 )" tls = "chain 'B' and (resid 561 through 585 )" tls = "chain 'B' and (resid 586 through 607 )" tls = "chain 'C' and (resid 461 through 470 )" tls = "chain 'C' and (resid 471 through 477 )" tls = "chain 'C' and (resid 478 through 489 )" tls = "chain 'C' and (resid 490 through 513 )" tls = "chain 'C' and (resid 514 through 560 )" tls = "chain 'C' and (resid 561 through 568 )" tls = "chain 'C' and (resid 569 through 583 )" tls = "chain 'C' and (resid 584 through 607 )" tls = "chain 'D' and (resid 461 through 477 )" tls = "chain 'D' and (resid 478 through 537 )" tls = "chain 'D' and (resid 538 through 607 )" tls = "chain 'E' and (resid 3 through 17 )" tls = "chain 'E' and (resid 18 through 42 )" tls = "chain 'E' and (resid 43 through 57 )" tls = "chain 'E' and (resid 58 through 59 )" tls = "chain 'F' and (resid 5 through 24 )" tls = "chain 'F' and (resid 25 through 44 )" tls = "chain 'F' and (resid 45 through 54 )" tls = "chain 'F' and (resid 55 through 57 )" tls = "chain 'I' and (resid 461 through 477 )" tls = "chain 'I' and (resid 478 through 513 )" tls = "chain 'I' and (resid 514 through 537 )" tls = "chain 'I' and (resid 538 through 560 )" tls = "chain 'I' and (resid 561 through 599 )" tls = "chain 'I' and (resid 600 through 607 )" tls = "chain 'J' and (resid 461 through 478 )" tls = "chain 'J' and (resid 479 through 504 )" tls = "chain 'J' and (resid 505 through 537 )" tls = "chain 'J' and (resid 538 through 547 )" tls = "chain 'J' and (resid 548 through 560 )" tls = "chain 'J' and (resid 561 through 585 )" tls = "chain 'J' and (resid 586 through 599 )" tls = "chain 'J' and (resid 600 through 607 )" tls = "chain 'K' and (resid 461 through 477 )" tls = "chain 'K' and (resid 478 through 491 )" tls = "chain 'K' and (resid 492 through 513 )" tls = "chain 'K' and (resid 514 through 560 )" tls = "chain 'K' and (resid 561 through 568 )" tls = "chain 'K' and (resid 569 through 599 )" tls = "chain 'K' and (resid 600 through 607 )" tls = "chain 'L' and (resid 461 through 537 )" tls = "chain 'L' and (resid 538 through 607 )" tls = "chain 'G' and (resid 8 through 32 )" tls = "chain 'G' and (resid 33 through 52 )" tls = "chain 'G' and (resid 53 through 56 )" tls = "chain 'H' and (resid 7 through 21 )" tls = "chain 'H' and (resid 22 through 46 )" tls = "chain 'H' and (resid 47 through 55 )"
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.01→40.9 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION
|
 Movie
Movie Controller
Controller









 PDBj
PDBj







































