[English] 日本語
 Yorodumi
Yorodumi- PDB-6o0h: Cryo-EM structure of human ATP-citrate lyase in complex with inhi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6o0h | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human ATP-citrate lyase in complex with inhibitor NDI-091143 | |||||||||||||||
 Components Components | ATP-citrate synthase | |||||||||||||||
 Keywords Keywords | ligase/ligase inhibitor / ATP-citrate lyase / LIGASE / ligase-ligase inhibitor complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationATP citrate synthase / ATP citrate synthase activity / citrate metabolic process / Fatty acyl-CoA biosynthesis / acetyl-CoA biosynthetic process / ChREBP activates metabolic gene expression / coenzyme A metabolic process / oxaloacetate metabolic process / negative regulation of ferroptosis / cholesterol biosynthetic process ...ATP citrate synthase / ATP citrate synthase activity / citrate metabolic process / Fatty acyl-CoA biosynthesis / acetyl-CoA biosynthetic process / ChREBP activates metabolic gene expression / coenzyme A metabolic process / oxaloacetate metabolic process / negative regulation of ferroptosis / cholesterol biosynthetic process / lipid biosynthetic process / fatty acid biosynthetic process / azurophil granule lumen / ficolin-1-rich granule lumen / ciliary basal body / Neutrophil degranulation / extracellular exosome / extracellular region / nucleoplasm / ATP binding / metal ion binding / membrane / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.67 Å | |||||||||||||||
 Authors Authors | Wei, J. / Tong, L. | |||||||||||||||
| Funding support |  United States, 4items United States, 4items
| |||||||||||||||
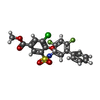
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: An allosteric mechanism for potent inhibition of human ATP-citrate lyase. Authors: Jia Wei / Silvana Leit / Jun Kuai / Eric Therrien / Salma Rafi / H James Harwood / Byron DeLaBarre / Liang Tong /  Abstract: ATP-citrate lyase (ACLY) is a central metabolic enzyme and catalyses the ATP-dependent conversion of citrate and coenzyme A (CoA) to oxaloacetate and acetyl-CoA. The acetyl-CoA product is crucial for ...ATP-citrate lyase (ACLY) is a central metabolic enzyme and catalyses the ATP-dependent conversion of citrate and coenzyme A (CoA) to oxaloacetate and acetyl-CoA. The acetyl-CoA product is crucial for the metabolism of fatty acids, the biosynthesis of cholesterol, and the acetylation and prenylation of proteins. There has been considerable interest in ACLY as a target for anti-cancer drugs, because many cancer cells depend on its activity for proliferation. ACLY is also a target against dyslipidaemia and hepatic steatosis, with a compound currently in phase 3 clinical trials. Many inhibitors of ACLY have been reported, but most of them have weak activity. Here we report the development of a series of low nanomolar, small-molecule inhibitors of human ACLY. We have also determined the structure of the full-length human ACLY homo-tetramer in complex with one of these inhibitors (NDI-091143) by cryo-electron microscopy, which reveals an unexpected mechanism of inhibition. The compound is located in an allosteric, mostly hydrophobic cavity next to the citrate-binding site, and requires extensive conformational changes in the enzyme that indirectly disrupt citrate binding. The observed binding mode is supported by and explains the structure-activity relationships of these compounds. This allosteric site greatly enhances the 'druggability' of ACLY and represents an attractive target for the development of new ACLY inhibitors. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6o0h.cif.gz 6o0h.cif.gz | 702.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6o0h.ent.gz pdb6o0h.ent.gz | 574.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6o0h.json.gz 6o0h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/o0/6o0h https://data.pdbj.org/pub/pdb/validation_reports/o0/6o0h ftp://data.pdbj.org/pub/pdb/validation_reports/o0/6o0h ftp://data.pdbj.org/pub/pdb/validation_reports/o0/6o0h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  0567MC  0568C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 122787.102 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ACLY / Production host: Homo sapiens (human) / Gene: ACLY / Production host:  #2: Chemical | ChemComp-LBG / #3: Chemical | ChemComp-ADP / |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: protein-inhibitor complex / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 4.7 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: C-flat-1.2/1.3 | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK II / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 130000 X / Nominal defocus max: -2000 nm / Nominal defocus min: -800 nm / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 10 sec. / Electron dose: 71 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 2803 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 250156 | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: D2 (2x2 fold dihedral) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.67 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 11452 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL |
 Movie
Movie Controller
Controller







 PDBj
PDBj