[English] 日本語
 Yorodumi
Yorodumi- PDB-6nvh: FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)ox... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6nvh | ||||||
|---|---|---|---|---|---|---|---|
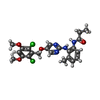
| Title | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | ||||||
 Components Components | Fibroblast growth factor receptor 4 | ||||||
 Keywords Keywords | TRANSFERASE/TRANSFERASE INHIBITOR / TRANSFERASE-TRANSFERASE INHIBITOR complex / TRANSFERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationFGFR4 mutant receptor activation / betaKlotho-mediated ligand binding / regulation of extracellular matrix disassembly / positive regulation of catalytic activity / phosphate ion homeostasis / regulation of bile acid biosynthetic process / FGFR4 ligand binding and activation / Phospholipase C-mediated cascade; FGFR4 / fibroblast growth factor receptor activity / positive regulation of DNA biosynthetic process ...FGFR4 mutant receptor activation / betaKlotho-mediated ligand binding / regulation of extracellular matrix disassembly / positive regulation of catalytic activity / phosphate ion homeostasis / regulation of bile acid biosynthetic process / FGFR4 ligand binding and activation / Phospholipase C-mediated cascade; FGFR4 / fibroblast growth factor receptor activity / positive regulation of DNA biosynthetic process / PI-3K cascade:FGFR4 / fibroblast growth factor binding / regulation of lipid metabolic process / positive regulation of proteolysis / PI3K Cascade / fibroblast growth factor receptor signaling pathway / SHC-mediated cascade:FGFR4 / transport vesicle / Signaling by FGFR4 in disease / FRS-mediated FGFR4 signaling / peptidyl-tyrosine phosphorylation / cholesterol homeostasis / Negative regulation of FGFR4 signaling / receptor protein-tyrosine kinase / Constitutive Signaling by Aberrant PI3K in Cancer / cell migration / PIP3 activates AKT signaling / glucose homeostasis / heparin binding / protein autophosphorylation / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / RAF/MAP kinase cascade / positive regulation of ERK1 and ERK2 cascade / receptor complex / endosome / positive regulation of cell population proliferation / positive regulation of gene expression / endoplasmic reticulum / Golgi apparatus / extracellular region / ATP binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Lin, X. / Smaill, J.B. / Squire, C.J. | ||||||
 Citation Citation |  Journal: Acs Med.Chem.Lett. / Year: 2019 Journal: Acs Med.Chem.Lett. / Year: 2019Title: Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4. Authors: Lin, X. / Yosaatmadja, Y. / Kalyukina, M. / Middleditch, M.J. / Zhang, Z. / Lu, X. / Ding, K. / Patterson, A.V. / Smaill, J.B. / Squire, C.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6nvh.cif.gz 6nvh.cif.gz | 72.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6nvh.ent.gz pdb6nvh.ent.gz | 51 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6nvh.json.gz 6nvh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nv/6nvh https://data.pdbj.org/pub/pdb/validation_reports/nv/6nvh ftp://data.pdbj.org/pub/pdb/validation_reports/nv/6nvh ftp://data.pdbj.org/pub/pdb/validation_reports/nv/6nvh | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6nvgC  6nviC  6nvjC  6nvkC  6nvlC  4xcuS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 33867.207 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FGFR4, JTK2, TKF / Production host: Homo sapiens (human) / Gene: FGFR4, JTK2, TKF / Production host:  References: UniProt: P22455, receptor protein-tyrosine kinase | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | | #3: Chemical | ChemComp-XL6 / | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.76 Å3/Da / Density % sol: 55.45 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop Details: 100mM Bis-tris pH 4.5, 200mM lithium sulphate, 12% PEG3350 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX2 / Wavelength: 0.9537 Å / Beamline: MX2 / Wavelength: 0.9537 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Aug 15, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9537 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→45.52 Å / Num. obs: 30981 / % possible obs: 100 % / Redundancy: 13.1 % / Biso Wilson estimate: 37.9 Å2 / CC1/2: 0.998 / Rpim(I) all: 0.023 / Net I/σ(I): 16.8 |
| Reflection shell | Resolution: 1.9→1.94 Å / Redundancy: 11.5 % / Mean I/σ(I) obs: 1.5 / Num. unique obs: 1946 / CC1/2: 0.623 / Rpim(I) all: 0.58 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4XCU Resolution: 1.9→45.52 Å / Cor.coef. Fo:Fc: 0.953 / Cor.coef. Fo:Fc free: 0.948 / SU B: 3.276 / SU ML: 0.093 / Cross valid method: THROUGHOUT / ESU R: 0.135 / ESU R Free: 0.124 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 39.829 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 1.9→45.52 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



















 PDBj
PDBj