[English] 日本語
 Yorodumi
Yorodumi- PDB-6lr2: SOLUTION STRUCTURE OF THE YTH DOMAIN IN YTH DOMAIN-2 CONTAINING P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6lr2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF THE YTH DOMAIN IN YTH DOMAIN-2 CONTAINING PROTEIN 2 | ||||||
 Components Components | YTH domain containing protein 2 (YTHDC2) | ||||||
 Keywords Keywords | RNA BINDING PROTEIN / MEIOSIS RELATED PROTEIN / Structural Genomics / PSI-2 / Protein Structure Initiative / RIKEN Structural Genomics/Proteomics Initiative / RSGI | ||||||
| Function / homology |  Function and homology information Function and homology informationgermline cell cycle switching, mitotic to meiotic cell cycle / ribonucleoprotein granule / host-mediated activation of viral genome replication / 3'-5' RNA helicase activity / N6-methyladenosine-containing RNA reader activity / oocyte development / ATP-dependent activity, acting on RNA / RNA polymerase binding / response to tumor necrosis factor / spermatid development ...germline cell cycle switching, mitotic to meiotic cell cycle / ribonucleoprotein granule / host-mediated activation of viral genome replication / 3'-5' RNA helicase activity / N6-methyladenosine-containing RNA reader activity / oocyte development / ATP-dependent activity, acting on RNA / RNA polymerase binding / response to tumor necrosis factor / spermatid development / response to interleukin-1 / meiotic cell cycle / helicase activity / RNA helicase / perinuclear region of cytoplasm / endoplasmic reticulum / ATP hydrolysis activity / RNA binding / ATP binding Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Muto, Y. / Kobayashi, N. / Yokoyama, S. / RIKEN Structural Genomics/Proteomics Initiative (RSGI) | ||||||
 Citation Citation |  Journal: TO BE PUBLISHED Journal: TO BE PUBLISHEDTitle: SOLUTION STRUCTURE OF THE YTH DOMAIN IN YTH DOMAIN-CONTAINING PROTEIN 2 Authors: Endo, R. / He, F. / Inoue, M. / Muto, Y. / Kigawa, T. / Shirouzu, M. / Yokoyama, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6lr2.cif.gz 6lr2.cif.gz | 869.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6lr2.ent.gz pdb6lr2.ent.gz | 729.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6lr2.json.gz 6lr2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6lr2_validation.pdf.gz 6lr2_validation.pdf.gz | 548.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6lr2_full_validation.pdf.gz 6lr2_full_validation.pdf.gz | 683.6 KB | Display | |
| Data in XML |  6lr2_validation.xml.gz 6lr2_validation.xml.gz | 49.6 KB | Display | |
| Data in CIF |  6lr2_validation.cif.gz 6lr2_validation.cif.gz | 81.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lr/6lr2 https://data.pdbj.org/pub/pdb/validation_reports/lr/6lr2 ftp://data.pdbj.org/pub/pdb/validation_reports/lr/6lr2 ftp://data.pdbj.org/pub/pdb/validation_reports/lr/6lr2 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 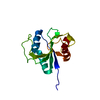
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 16138.065 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: P060508-16 Homo sapiens (human) / Plasmid: P060508-16Production host: Cell-free gateway cloning vector N-term 8xHis eGFP pCellFree_G03 (others) References: UniProt: Q6ZMY0, UniProt: Q9H6S0*PLUS |
|---|---|
| Sequence details | Authors state that 6L2R has a mutation, L129Q based rs1132529 in dbSNP database. |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Sample state: isotropic / Type: 3D 13C,15N-SEPARATED NOESY SPECTRA |
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 1.0 mg/mL [U-99% 13C; U-99% 15N] helicase, 90% H2O/10% D2O Details: 1mM D-DTT;0.02% NaN3 were used to keep the protein condition Label: 13C, 15N_sample / Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 1.0 mg/mL / Component: helicase / Isotopic labeling: [U-99% 13C; U-99% 15N] |
| Sample conditions | Details: 20mM D-Tris-HCL (PH7.0); 100mM NaCl; 1mM D-DTT;0.02% NaN3 Ionic strength: 100 mM / Label: condition_1 / pH: 7 / Pressure: 1 atm / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 800 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: the structures are based on 3191 NOE-derived distance contrrains, 71 main chain dihedral angle constraints based on TALOS program and 51 side chain dihedral constraints based on NOE pattern. | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with favorable non-bond energy Conformers calculated total number: 200 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller











 PDBj
PDBj
 gel filtration
gel filtration