+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6lkn | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of ATP11C-CDC50A in PtdSer-bound E2P state | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / P-type ATPase / flippase / apoptosis / phosphatidylserine | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of phospholipid translocation / aminophospholipid flippase activity / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / positive regulation of protein exit from endoplasmic reticulum / ATPase-coupled intramembrane lipid transporter activity / phosphatidylethanolamine flippase activity ...positive regulation of phospholipid translocation / aminophospholipid flippase activity / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / positive regulation of protein exit from endoplasmic reticulum / ATPase-coupled intramembrane lipid transporter activity / phosphatidylethanolamine flippase activity / xenobiotic transmembrane transport / P-type phospholipid transporter / phospholipid translocation / azurophil granule membrane / transport vesicle membrane / Ion transport by P-type ATPases / specific granule membrane / positive regulation of neuron projection development / recycling endosome / recycling endosome membrane / late endosome membrane / early endosome membrane / monoatomic ion transmembrane transport / apical plasma membrane / lysosomal membrane / Neutrophil degranulation / endoplasmic reticulum membrane / structural molecule activity / magnesium ion binding / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.9 Å MOLECULAR REPLACEMENT / Resolution: 3.9 Å | |||||||||
 Authors Authors | Abe, K. / Irie, K. / Nakanishi, H. / Hasegawa, K. | |||||||||
| Funding support |  Japan, 2items Japan, 2items
| |||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2020 Journal: J.Biol.Chem. / Year: 2020Title: Crystal structure of a human plasma membrane phospholipid flippase. Authors: Nakanishi, H. / Irie, K. / Segawa, K. / Hasegawa, K. / Fujiyoshi, Y. / Nagata, S. / Abe, K. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6lkn.cif.gz 6lkn.cif.gz | 2.5 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6lkn.ent.gz pdb6lkn.ent.gz | 1.9 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6lkn.json.gz 6lkn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lk/6lkn https://data.pdbj.org/pub/pdb/validation_reports/lk/6lkn ftp://data.pdbj.org/pub/pdb/validation_reports/lk/6lkn ftp://data.pdbj.org/pub/pdb/validation_reports/lk/6lkn | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6k7lS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| 4 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 8 molecules AEIMCFJN
| #1: Protein | Mass: 129710.633 Da / Num. of mol.: 4 / Mutation: C111W Source method: isolated from a genetically manipulated source Details: Amino acid 410 in PDB is BFD, which is BeF-bound Asp. Source: (gene. exp.)  Homo sapiens (human) / Gene: ATP11C, ATPIG, ATPIQ / Cell line (production host): Expi293 / Production host: Homo sapiens (human) / Gene: ATP11C, ATPIG, ATPIQ / Cell line (production host): Expi293 / Production host:  Homo sapiens (human) Homo sapiens (human)References: UniProt: Q8NB49, P-type phospholipid transporter #2: Protein | Mass: 40840.684 Da / Num. of mol.: 4 / Mutation: N190Q, S292W Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TMEM30A, C6orf67, CDC50A / Cell line (production host): Expi293 / Production host: Homo sapiens (human) / Gene: TMEM30A, C6orf67, CDC50A / Cell line (production host): Expi293 / Production host:  Homo sapiens (human) / References: UniProt: Q9NV96 Homo sapiens (human) / References: UniProt: Q9NV96 |
|---|
-Sugars , 2 types, 20 molecules 
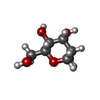

| #5: Sugar | ChemComp-NAG / #6: Sugar | ChemComp-AH2 / |
|---|
-Non-polymers , 3 types, 19 molecules 




| #3: Chemical | ChemComp-P5S / #4: Chemical | ChemComp-MG / #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.23 Å3/Da / Density % sol: 70.9 % |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 14-15% PEG4000, 10% glycerol, 5 mM beta-mercaptoethanol, 0.4 M MgSO4 |
-Data collection
| Diffraction | Mean temperature: 80 K / Ambient temp details: LN2 / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 1 Å / Beamline: BL41XU / Wavelength: 1 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Apr 19, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3.9→50 Å / Num. obs: 106458 / % possible obs: 71.32 % / Redundancy: 776.9 % / Biso Wilson estimate: 98.24 Å2 / CC1/2: 0.92 / Rmerge(I) obs: 1.141 / Rpim(I) all: 0.0419 / Net I/σ(I): 15.88 |
| Reflection shell | Resolution: 3.9→4 Å / Redundancy: 698.5 % / Num. unique obs: 1223 / CC1/2: 0.864 / % possible all: 11.64 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6k7l Resolution: 3.9→49.97 Å / SU ML: 0.6547 / Cross valid method: FREE R-VALUE / σ(F): 2.07 / Phase error: 38.7776 Details: Diffractions were strongly anisorlopic. We merged 1588 crystal data to improve data quality.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 161.97 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.9→49.97 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -25.8822080037 Å / Origin y: 100.493022409 Å / Origin z: -125.137573873 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: all |
 Movie
Movie Controller
Controller













 PDBj
PDBj









