[English] 日本語
 Yorodumi
Yorodumi- PDB-6k0r: Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphoryl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6k0r | ||||||
|---|---|---|---|---|---|---|---|
| Title | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | ||||||
 Components Components |
| ||||||
 Keywords Keywords | CIRCADIAN CLOCK PROTEIN / ATPase / Cordycepin | ||||||
| Function / homology |  Function and homology information Function and homology informationpromoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / establishment of protein localization to chromatin / R2TP complex / dynein axonemal particle / RPAP3/R2TP/prefoldin-like complex / Swr1 complex / Ino80 complex ...promoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / establishment of protein localization to chromatin / R2TP complex / dynein axonemal particle / RPAP3/R2TP/prefoldin-like complex / Swr1 complex / Ino80 complex / regulation of double-strand break repair / box C/D snoRNP assembly / regulation of chromosome organization / NuA4 histone acetyltransferase complex / TFIID-class transcription factor complex binding / regulation of DNA replication / MLL1 complex / regulation of embryonic development / Telomere Extension By Telomerase / protein folding chaperone complex / RNA polymerase II core promoter sequence-specific DNA binding / regulation of DNA repair / positive regulation of double-strand break repair via homologous recombination / Deposition of new CENPA-containing nucleosomes at the centromere / TBP-class protein binding / DNA helicase activity / telomere maintenance / positive regulation of DNA repair / cellular response to estradiol stimulus / euchromatin / negative regulation of canonical Wnt signaling pathway / Formation of the beta-catenin:TCF transactivating complex / ADP binding / beta-catenin binding / chromatin DNA binding / DNA Damage Recognition in GG-NER / nuclear matrix / cellular response to UV / unfolded protein binding / UCH proteinases / transcription corepressor activity / positive regulation of canonical Wnt signaling pathway / nucleosome / protein folding / HATs acetylate histones / ATPase binding / regulation of apoptotic process / DNA recombination / spermatogenesis / DNA helicase / transcription coactivator activity / regulation of cell cycle / Ub-specific processing proteases / protein stabilization / nuclear speck / ciliary basal body / cadherin binding / RNA polymerase II cis-regulatory region sequence-specific DNA binding / chromatin remodeling / ribonucleoprotein complex / cell division / DNA repair / centrosome / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / extracellular exosome / nucleoplasm / ATP binding / identical protein binding / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.502 Å MOLECULAR REPLACEMENT / Resolution: 2.502 Å | ||||||
 Authors Authors | Zhang, W. / Chen, L. / Li, W. / Ju, D. / Huang, N. / Zhang, E. | ||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2020 Journal: Sci Transl Med / Year: 2020Title: Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals. Authors: Ju, D. / Zhang, W. / Yan, J. / Zhao, H. / Li, W. / Wang, J. / Liao, M. / Xu, Z. / Wang, Z. / Zhou, G. / Mei, L. / Hou, N. / Ying, S. / Cai, T. / Chen, S. / Xie, X. / Lai, L. / Tang, C. / ...Authors: Ju, D. / Zhang, W. / Yan, J. / Zhao, H. / Li, W. / Wang, J. / Liao, M. / Xu, Z. / Wang, Z. / Zhou, G. / Mei, L. / Hou, N. / Ying, S. / Cai, T. / Chen, S. / Xie, X. / Lai, L. / Tang, C. / Park, N. / Takahashi, J.S. / Huang, N. / Qi, X. / Zhang, E.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6k0r.cif.gz 6k0r.cif.gz | 694.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6k0r.ent.gz pdb6k0r.ent.gz | 554.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6k0r.json.gz 6k0r.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k0/6k0r https://data.pdbj.org/pub/pdb/validation_reports/k0/6k0r ftp://data.pdbj.org/pub/pdb/validation_reports/k0/6k0r ftp://data.pdbj.org/pub/pdb/validation_reports/k0/6k0r | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2xszS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 12 molecules ABCGHIDEFJKL
| #1: Protein | Mass: 38840.000 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: RUVBL1, INO80H, NMP238, TIP49, TIP49A / Production host: Homo sapiens (human) / Gene: RUVBL1, INO80H, NMP238, TIP49, TIP49A / Production host:  #2: Protein | Mass: 40263.965 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: RUVBL2, INO80J, TIP48, TIP49B, CGI-46 / Production host: Homo sapiens (human) / Gene: RUVBL2, INO80J, TIP48, TIP49B, CGI-46 / Production host:  |
|---|
-Non-polymers , 6 types, 20 molecules 


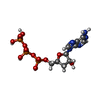







| #3: Chemical | ChemComp-ADP / | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #4: Chemical | ChemComp-CUU / [( #5: Chemical | #6: Chemical | #7: Chemical | ChemComp-CU0 / [( | #8: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.55 Å3/Da / Density % sol: 51.71 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop Details: 0.1 M HEPES-Na pH 7.5-7.6, 0.2 M MgCl2, 20-21 % PEG400 PH range: 7.5-7.6 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL17U1 / Wavelength: 0.97918 Å / Beamline: BL17U1 / Wavelength: 0.97918 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Mar 16, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97918 Å / Relative weight: 1 |
| Reflection | Resolution: 2.502→235.137 Å / Num. obs: 90883 / % possible obs: 89.9 % / Redundancy: 4.6 % / Biso Wilson estimate: 62.45 Å2 / CC1/2: 0.996 / Rmerge(I) obs: 0.107 / Rpim(I) all: 0.054 / Rrim(I) all: 0.12 / Net I/σ(I): 9.1 |
| Reflection shell | Resolution: 2.502→2.81 Å / Rmerge(I) obs: 1.033 / Num. unique obs: 4542 / CC1/2: 0.159 / Rpim(I) all: 0.474 / Rrim(I) all: 1.138 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2XSZ Resolution: 2.502→49.72 Å / Cor.coef. Fo:Fc: 0.891 / Cor.coef. Fo:Fc free: 0.847 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.425
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 174.13 Å2 / Biso mean: 70.63 Å2 / Biso min: 26.19 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.502→49.72 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.502→2.68 Å / Rfactor Rfree error: 0 / Total num. of bins used: 50
|
 Movie
Movie Controller
Controller











 PDBj
PDBj









