| 登録情報 | データベース: PDB / ID: 6jpv
|
|---|
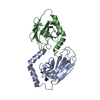
| タイトル | Structural analysis of AIMP2-DX2 and HSP70 interaction |
|---|
 要素 要素 | Heat shock 70 kDa protein 1A,Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 |
|---|
 キーワード キーワード | CHAPERONE / HSP70 / AIMP2-DX2 / substrate binding domain |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
type II pneumocyte differentiation / : / Selenoamino acid metabolism / denatured protein binding / cellular heat acclimation / negative regulation of inclusion body assembly / Viral RNP Complexes in the Host Cell Nucleus / death receptor agonist activity / C3HC4-type RING finger domain binding / positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway ...type II pneumocyte differentiation / : / Selenoamino acid metabolism / denatured protein binding / cellular heat acclimation / negative regulation of inclusion body assembly / Viral RNP Complexes in the Host Cell Nucleus / death receptor agonist activity / C3HC4-type RING finger domain binding / positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway / Cytosolic tRNA aminoacylation / aminoacyl-tRNA synthetase multienzyme complex / positive regulation of microtubule nucleation / ATP-dependent protein disaggregase activity / misfolded protein binding / negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / positive regulation of tumor necrosis factor-mediated signaling pathway / regulation of mitotic spindle assembly / aggresome / lysosomal transport / cellular response to steroid hormone stimulus / mRNA catabolic process / : / regulation of protein ubiquitination / Regulation of HSF1-mediated heat shock response / HSF1-dependent transactivation / cellular response to unfolded protein / response to unfolded protein / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / Mitochondrial unfolded protein response (UPRmt) / Attenuation phase / chaperone-mediated protein complex assembly / ATP metabolic process / negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway / transcription regulator inhibitor activity / heat shock protein binding / inclusion body / negative regulation of protein ubiquitination / protein folding chaperone / centriole / Transcriptional and post-translational regulation of MITF-M expression and activity / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / positive regulation of erythrocyte differentiation / positive regulation of RNA splicing / positive regulation of protein ubiquitination / positive regulation of interleukin-8 production / AUF1 (hnRNP D0) binds and destabilizes mRNA / ATP-dependent protein folding chaperone / negative regulation of transforming growth factor beta receptor signaling pathway / G protein-coupled receptor binding / positive regulation of NF-kappaB transcription factor activity / negative regulation of cell growth / PKR-mediated signaling / histone deacetylase binding / disordered domain specific binding / transcription corepressor activity / unfolded protein binding / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / positive regulation of neuron apoptotic process / cellular response to heat / virus receptor activity / protein refolding / cellular response to oxidative stress / protein-containing complex assembly / blood microparticle / molecular adaptor activity / vesicle / ficolin-1-rich granule lumen / protein stabilization / protein ubiquitination / nuclear speck / cadherin binding / translation / receptor ligand activity / ribonucleoprotein complex / signaling receptor binding / negative regulation of cell population proliferation / focal adhesion / apoptotic process / ubiquitin protein ligase binding / centrosome / Neutrophil degranulation / positive regulation of gene expression / negative regulation of apoptotic process / perinuclear region of cytoplasm / enzyme binding / endoplasmic reticulum / negative regulation of transcription by RNA polymerase II / protein-containing complex / ATP hydrolysis activity / mitochondrion / extracellular space / RNA binding / extracellular exosome / extracellular region / nucleoplasm / ATP binding / nucleus / membrane / plasma membrane類似検索 - 分子機能 AIMP2, lysyl-tRNA synthetase binding domain / AIMP2, thioredoxin-like domain / Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 / AIMP2 lysyl-tRNA synthetase binding domain / Thioredoxin-like domain / Substrate Binding Domain Of DNAk; Chain A, domain 1 / Substrate Binding Domain Of DNAk; Chain A, domain 1 / Heat shock hsp70 proteins family signature 2. / Heat shock hsp70 proteins family signature 1. / Heat shock hsp70 proteins family signature 3. ...AIMP2, lysyl-tRNA synthetase binding domain / AIMP2, thioredoxin-like domain / Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 / AIMP2 lysyl-tRNA synthetase binding domain / Thioredoxin-like domain / Substrate Binding Domain Of DNAk; Chain A, domain 1 / Substrate Binding Domain Of DNAk; Chain A, domain 1 / Heat shock hsp70 proteins family signature 2. / Heat shock hsp70 proteins family signature 1. / Heat shock hsp70 proteins family signature 3. / Heat shock protein 70, conserved site / Glutathione S-transferase, C-terminal domain / Heat shock protein 70kD, peptide-binding domain superfamily / Heat shock protein 70 family / Hsp70 protein / Heat shock protein 70kD, C-terminal domain superfamily / Glutathione S-transferase, C-terminal / Glutathione S-transferase, C-terminal domain superfamily / ATPase, nucleotide binding domain / Sandwich / Mainly Beta類似検索 - ドメイン・相同性 Heat shock 70 kDa protein 1A / Aminoacyl tRNA synthase complex-interacting multifunctional protein 2類似検索 - 構成要素 |
|---|
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.15000649024 Å 分子置換 / 解像度: 2.15000649024 Å |
|---|
 データ登録者 データ登録者 | Cho, H.Y. / Son, S.Y. / Jeon, Y.H. |
|---|
| 資金援助 |  韓国, 1件 韓国, 1件 | 組織 | 認可番号 | 国 |
|---|
| National Research Foundation (Korea) | NRF-2013M3A6A4045160 |  韓国 韓国 |
|
|---|
 引用 引用 |  ジャーナル: Nat.Chem.Biol. / 年: 2020 ジャーナル: Nat.Chem.Biol. / 年: 2020
タイトル: Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development.
著者: Lim, S. / Cho, H.Y. / Kim, D.G. / Roh, Y. / Son, S.Y. / Mushtaq, A.U. / Kim, M. / Bhattarai, D. / Sivaraman, A. / Lee, Y. / Lee, J. / Yang, W.S. / Kim, H.K. / Kim, M.H. / Lee, K. / Jeon, Y.H. / Kim, S. |
|---|
| 履歴 | | 登録 | 2019年3月28日 | 登録サイト: PDBJ / 処理サイト: PDBJ |
|---|
| 改定 1.0 | 2019年10月2日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2019年11月20日 | Group: Database references / カテゴリ: citation / Item: _citation.pdbx_database_id_DOI / _citation.title |
|---|
| 改定 1.2 | 2019年12月18日 | Group: Database references / カテゴリ: citation / citation_author / Item: _citation.pdbx_database_id_PubMed |
|---|
| 改定 1.3 | 2020年1月1日 | Group: Database references / カテゴリ: citation / citation_author
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation.year / _citation_author.identifier_ORCID |
|---|
| 改定 1.4 | 2023年11月22日 | Group: Data collection / Database references / Refinement description
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.15000649024 Å
分子置換 / 解像度: 2.15000649024 Å  データ登録者
データ登録者 韓国, 1件
韓国, 1件  引用
引用 ジャーナル: Nat.Chem.Biol. / 年: 2020
ジャーナル: Nat.Chem.Biol. / 年: 2020 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 6jpv.cif.gz
6jpv.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb6jpv.ent.gz
pdb6jpv.ent.gz PDB形式
PDB形式 6jpv.json.gz
6jpv.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 6jpv_validation.pdf.gz
6jpv_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 6jpv_full_validation.pdf.gz
6jpv_full_validation.pdf.gz 6jpv_validation.xml.gz
6jpv_validation.xml.gz 6jpv_validation.cif.gz
6jpv_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/jp/6jpv
https://data.pdbj.org/pub/pdb/validation_reports/jp/6jpv ftp://data.pdbj.org/pub/pdb/validation_reports/jp/6jpv
ftp://data.pdbj.org/pub/pdb/validation_reports/jp/6jpv リンク
リンク 集合体
集合体
 要素
要素 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト: PAL/PLS
シンクロトロン / サイト: PAL/PLS  / ビームライン: 5C (4A) / 波長: 0.9795 Å
/ ビームライン: 5C (4A) / 波長: 0.9795 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー















 PDBj
PDBj










