[English] 日本語
 Yorodumi
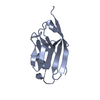
Yorodumi- PDB-6gcj: Solution structure of the RodA hydrophobin from Aspergillus fumigatus -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6gcj | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Solution structure of the RodA hydrophobin from Aspergillus fumigatus | |||||||||
 Components Components | Hydrophobin | |||||||||
 Keywords Keywords | STRUCTURAL PROTEIN / Soluble form of the protein that coats Aspergillus fumigatus conidia | |||||||||
| Function / homology | Fungal hydrophobin / Hydrophobin, conserved site / Fungal hydrophobins signature. / Hydrophobin / Hydrophobins / structural constituent of cell wall / fungal-type cell wall / extracellular region / Hydrophobin Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | SOLUTION NMR / simulated annealing | |||||||||
 Authors Authors | Pille, A. / Kwan, A. / Aimanianda, V. / Latge, J.-P. / Sunde, M. / Guijarro, J.I. | |||||||||
| Funding support |  France, 2items France, 2items
| |||||||||
 Citation Citation |  Journal: Cell Surf / Year: 2019 Journal: Cell Surf / Year: 2019Title: Assembly and disassembly of Aspergillus fumigatus conidial rodlets Authors: Valsecchi, I. / Lai, J. / Emmanuel, S.-V. / Pille, A. / Beaussart, A. / Lo, V. / Chi, L. / Pham, L. / Kwan, A.H. / Matondo, M. / Duchateau, M. / Giai-Giannetto, Q. / Aimanianda, V. / ...Authors: Valsecchi, I. / Lai, J. / Emmanuel, S.-V. / Pille, A. / Beaussart, A. / Lo, V. / Chi, L. / Pham, L. / Kwan, A.H. / Matondo, M. / Duchateau, M. / Giai-Giannetto, Q. / Aimanianda, V. / Dufrene, Y. / Bayry, J. / Guijarro, J.I. / Sunde, M. / Latge, J.P. #1: Journal: Biomol NMR Assign / Year: 2015 Title: (1)H, (13)C and (15)N resonance assignments of the RodA hydrophobin from the opportunistic pathogen Aspergillus fumigatus. Authors: Pille, A. / Kwan, A.H. / Cheung, I. / Hampsey, M. / Aimanianda, V. / Delepierre, M. / Latge, J.P. / Sunde, M. / Guijarro, J.I. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6gcj.cif.gz 6gcj.cif.gz | 348.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6gcj.ent.gz pdb6gcj.ent.gz | 286.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6gcj.json.gz 6gcj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gc/6gcj https://data.pdbj.org/pub/pdb/validation_reports/gc/6gcj ftp://data.pdbj.org/pub/pdb/validation_reports/gc/6gcj ftp://data.pdbj.org/pub/pdb/validation_reports/gc/6gcj | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 14502.397 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: NMR EXPERIMENTS AND STRUCTURE CALCULATIONS WERE PERFORMED WITH THE PREDICTED FULL-LENGTH PROTEIN (RESIDUES 19-159) AFTER CLEAVAGE OF THE N-TERMINAL SECRETION SIGNAL PEPTIDE (1-18). BECAUSE ...Details: NMR EXPERIMENTS AND STRUCTURE CALCULATIONS WERE PERFORMED WITH THE PREDICTED FULL-LENGTH PROTEIN (RESIDUES 19-159) AFTER CLEAVAGE OF THE N-TERMINAL SECRETION SIGNAL PEPTIDE (1-18). BECAUSE RESIDUES 19-39 ARE DISORDERED, ONLY THE STRUCTURE FOR RESIDUES 39-159 IS DEPOSITED. Source: (gene. exp.)  Strain: CEA10 / CBS 144.89 / FGSC A1163 / Gene: AFUB_057130 / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 0.36 mM [U-99% 13C; U-99% 15N] RodA, 90% H2O/10% D2O Label: 15N_13C_Sample / Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 0.36 mM / Component: RodA / Isotopic labeling: [U-99% 13C; U-99% 15N] |
| Sample conditions | Ionic strength: 20 mM / Ionic strength err: 0.1 / Label: conditions_1 / pH: 4.3 / PH err: 0.05 / Pressure: 1 atm / Temperature: 298.15 K / Temperature err: 0.1 |
-NMR measurement
| NMR spectrometer | Type: Agilent NMR System / Manufacturer: Agilent / Model: NMR System / Field strength: 600 MHz / Details: Equipped with a cryoprobe |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 5 Details: The structures are based on a total of 2737 restraints, 1590 are unambiguous and 987 ambiguous NOE-derived distances restraints and from 160 dihedral angle constraints derived from Talos-N and HNHA experiment. | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 500 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller











 PDBj
PDBj HSQC
HSQC