+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6eg2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human BRM in complex with compound 16 | ||||||
 Components Components | Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | ||||||
 Keywords Keywords | Hydrolase/Hydrolase Inhibitor / Helicase / ATPase / Chromatin remodeling / inhibitor / MBP fusion / Hydrolase-Hydrolase Inhibitor complex | ||||||
| Function / homology |  Function and homology information Function and homology informationbBAF complex / npBAF complex / nBAF complex / brahma complex / nucleosome array spacer activity / GBAF complex / regulation of G0 to G1 transition / intermediate filament cytoskeleton / regulation of nucleotide-excision repair / regulation of mitotic metaphase/anaphase transition ...bBAF complex / npBAF complex / nBAF complex / brahma complex / nucleosome array spacer activity / GBAF complex / regulation of G0 to G1 transition / intermediate filament cytoskeleton / regulation of nucleotide-excision repair / regulation of mitotic metaphase/anaphase transition / SWI/SNF complex / positive regulation of T cell differentiation / positive regulation of double-strand break repair / positive regulation of stem cell population maintenance / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / Regulation of MITF-M-dependent genes involved in pigmentation / regulation of G1/S transition of mitotic cell cycle / carbohydrate transmembrane transporter activity / maltose binding / negative regulation of cell differentiation / spermatid development / maltose transport / maltodextrin transmembrane transport / positive regulation of myoblast differentiation / ATP-dependent activity, acting on DNA / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / positive regulation of cell differentiation / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / helicase activity / negative regulation of cell growth / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / RMTs methylate histone arginines / nervous system development / outer membrane-bounded periplasmic space / histone binding / transcription coactivator activity / transcription cis-regulatory region binding / chromatin remodeling / intracellular membrane-bounded organelle / negative regulation of cell population proliferation / hydrolase activity / negative regulation of DNA-templated transcription / positive regulation of cell population proliferation / chromatin binding / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.98 Å MOLECULAR REPLACEMENT / Resolution: 2.98 Å | ||||||
 Authors Authors | Zhu, X. / Kulathila, R. / Hu, T. / Xie, X. | ||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2018 Journal: J. Med. Chem. / Year: 2018Title: Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. Authors: Papillon, J.P.N. / Nakajima, K. / Adair, C.D. / Hempel, J. / Jouk, A.O. / Karki, R.G. / Mathieu, S. / Mobitz, H. / Ntaganda, R. / Smith, T. / Visser, M. / Hill, S.E. / Hurtado, F.K. / ...Authors: Papillon, J.P.N. / Nakajima, K. / Adair, C.D. / Hempel, J. / Jouk, A.O. / Karki, R.G. / Mathieu, S. / Mobitz, H. / Ntaganda, R. / Smith, T. / Visser, M. / Hill, S.E. / Hurtado, F.K. / Chenail, G. / Bhang, H.C. / Bric, A. / Xiang, K. / Bushold, G. / Gilbert, T. / Vattay, A. / Dooley, J. / Costa, E.A. / Park, I. / Li, A. / Farley, D. / Lounkine, E. / Yue, Q.K. / Xie, X. / Zhu, X. / Kulathila, R. / King, D. / Hu, T. / Vulic, K. / Cantwell, J. / Luu, C. / Jagani, Z. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6eg2.cif.gz 6eg2.cif.gz | 138 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6eg2.ent.gz pdb6eg2.ent.gz | 105.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6eg2.json.gz 6eg2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eg/6eg2 https://data.pdbj.org/pub/pdb/validation_reports/eg/6eg2 ftp://data.pdbj.org/pub/pdb/validation_reports/eg/6eg2 ftp://data.pdbj.org/pub/pdb/validation_reports/eg/6eg2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6eg3C  1fqaS  5hzrS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 69653.055 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human)Gene: malE, Z5632, ECs5017, SMARCA2, BAF190B, BRM, SNF2A, SNF2L2 Production host: Escherichia coli References: UniProt: P0AEY0, UniProt: P51531, Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement | ||
|---|---|---|---|
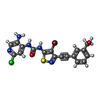
| #2: Chemical | ChemComp-J7J / | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.05 Å3/Da / Density % sol: 69.65 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: 100mM Hepes pH7.5, 200mM sodium chloride, 8% isopropanol |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 Å / Beamline: 17-ID / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Nov 20, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.98→120.688 Å / Num. obs: 23436 / % possible obs: 97.9 % / Redundancy: 12.7 % / Net I/σ(I): 18.2 |
| Reflection shell | Resolution: 2.98→2.99 Å |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1FQA, 5HZR Resolution: 2.98→96.73 Å / Cor.coef. Fo:Fc: 0.8518 / Cor.coef. Fo:Fc free: 0.8471 / SU R Cruickshank DPI: 0.874 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.851 / SU Rfree Blow DPI: 0.41 / SU Rfree Cruickshank DPI: 0.418
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 189.41 Å2 / Biso mean: 102.83 Å2 / Biso min: 36.11 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.552 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.98→96.73 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.98→3.11 Å / Rfactor Rfree error: 0 / Total num. of bins used: 12
|
 Movie
Movie Controller
Controller











 PDBj
PDBj