+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6.0E+20 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Dario rerio galectin-1-L2 | |||||||||
 Components Components | Galectin | |||||||||
 Keywords Keywords | SUGAR BINDING PROTEIN / Galectin-1 / innate immunity infectious hematopoietic necrosis virus (IHNV) / Danio rerio / zebrafish | |||||||||
| Function / homology |  Function and homology information Function and homology informationgalactoside binding / vasculature development / sprouting angiogenesis / positive regulation of neurogenesis / positive regulation of wound healing / skeletal muscle fiber development / laminin binding / carbohydrate binding / extracellular space / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | |||||||||
 Authors Authors | Ghosh, A. / Bianchet, M.A. | |||||||||
| Funding support |  United States, 1items United States, 1items
| |||||||||
 Citation Citation |  Journal: Glycobiology / Year: 2019 Journal: Glycobiology / Year: 2019Title: Structure of the zebrafish galectin-1-L2 and model of its interaction with the infectious hematopoietic necrosis virus (IHNV) envelope glycoprotein. Authors: Ghosh, A. / Banerjee, A. / Amzel, L.M. / Vasta, G.R. / Bianchet, M.A. #1:  Journal: Proteins / Year: 2000 Journal: Proteins / Year: 2000Title: Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes. Authors: Bianchet, M.A. / Ahmed, H. / Vasta, G.R. / Amzel, L.M. #2: Journal: J. Biol. Chem. / Year: 2013 Title: The galectin CvGal1 from the eastern oyster (Crassostrea virginica) binds to blood group A oligosaccharides on the hemocyte surface. Authors: Feng, C. / Ghosh, A. / Amin, M.N. / Giomarelli, B. / Shridhar, S. / Banerjee, A. / Fernandez-Robledo, J.A. / Bianchet, M.A. / Wang, L.X. / Wilson, I.B. / Vasta, G.R. #3: Journal: Biochemistry / Year: 2015 Title: Galectin CvGal2 from the Eastern Oyster (Crassostrea virginica) Displays Unique Specificity for ABH Blood Group Oligosaccharides and Differentially Recognizes Sympatric Perkinsus Species. Authors: Feng, C. / Ghosh, A. / Amin, M.N. / Bachvaroff, T.R. / Tasumi, S. / Pasek, M. / Banerjee, A. / Shridhar, S. / Wang, L.X. / Bianchet, M.A. / Vasta, G.R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6e20.cif.gz 6e20.cif.gz | 75.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6e20.ent.gz pdb6e20.ent.gz | 54 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6e20.json.gz 6e20.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6e20_validation.pdf.gz 6e20_validation.pdf.gz | 1.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6e20_full_validation.pdf.gz 6e20_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  6e20_validation.xml.gz 6e20_validation.xml.gz | 14.6 KB | Display | |
| Data in CIF |  6e20_validation.cif.gz 6e20_validation.cif.gz | 19.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e2/6e20 https://data.pdbj.org/pub/pdb/validation_reports/e2/6e20 ftp://data.pdbj.org/pub/pdb/validation_reports/e2/6e20 ftp://data.pdbj.org/pub/pdb/validation_reports/e2/6e20 | HTTPS FTP |
-Related structure data
| Related structure data | 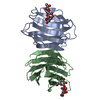 1ganS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 15421.234 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Polysaccharide | #3: Chemical | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.32 Å3/Da / Density % sol: 47.07 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: 0.1 M HEPES pH 7.5, 0.2 M Magnesium Chloride Hexahydrate, 30% (v/v) PolyethG 400 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.5 Å ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.5 Å |
| Detector | Type: RIGAKU SATURN 944 / Detector: CCD / Date: Aug 22, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5 Å / Relative weight: 1 |
| Reflection | Resolution: 2→35 Å / Num. obs: 18718 / % possible obs: 99.2 % / Redundancy: 3.5 % / Rmerge(I) obs: 0.046 / Net I/σ(I): 33.5 |
| Reflection shell | Resolution: 2→2.05 Å / Redundancy: 2.7 % / Rmerge(I) obs: 0.218 / Mean I/σ(I) obs: 6.5 / Num. unique obs: 1262 / % possible all: 97.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1GAN Resolution: 2→34.81 Å / Cor.coef. Fo:Fc: 0.96 / Cor.coef. Fo:Fc free: 0.936 / SU B: 4.772 / SU ML: 0.131 / Cross valid method: THROUGHOUT / ESU R: 0.189 / ESU R Free: 0.174 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 29.813 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2→34.81 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj