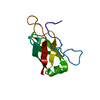
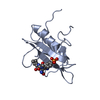
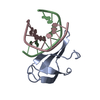
Entry Database : PDB / ID : 6d7yTitle 1.75 Angstrom Resolution Crystal Structure of the Toxic C-Terminal Tip of CdiA from Pseudomonas aeruginosa in Complex with Immune Protein Hemagglutinin immune protein Keywords / / / Function / homology / Biological species Pseudomonas aeruginosa (bacteria)Enterobacter cloacae (bacteria)Method / / / Resolution : 1.75 Å Authors Minasov, G. / Shuvalova, L. / Wawrzak, Z. / Kiryukhina, O. / Allen, J.P. / Hauser, A.R. / Anderson, W.F. / Satchell, K.J.F. / Joachimiak, A. / Center for Structural Genomics of Infectious Diseases (CSGID) Journal : Proc.Natl.Acad.Sci.USA / Year : 2020Title : A comparative genomics approach identifies contact-dependent growth inhibition as a virulence determinant.Authors : Allen, J.P. / Ozer, E.A. / Minasov, G. / Shuvalova, L. / Kiryukhina, O. / Satchell, K.J.F. / Hauser, A.R. History Deposition Apr 25, 2018 Deposition site / Processing site Revision 1.0 May 1, 2019 Provider / Type Revision 1.1 Mar 11, 2020 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.title / _citation.year Revision 1.2 Mar 25, 2020 Group / Category / citation_authorItem / _citation.title / _citation_author.nameRevision 1.3 Apr 15, 2020 Group / Category Item / _citation.page_first / _citation.page_lastRevision 1.4 Oct 16, 2024 Group / Database references / Structure summaryCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 Enterobacter cloacae (bacteria)
Enterobacter cloacae (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SAD / Resolution: 1.75 Å
SAD / Resolution: 1.75 Å  Authors
Authors Citation
Citation Journal: Proc.Natl.Acad.Sci.USA / Year: 2020
Journal: Proc.Natl.Acad.Sci.USA / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6d7y.cif.gz
6d7y.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6d7y.ent.gz
pdb6d7y.ent.gz PDB format
PDB format 6d7y.json.gz
6d7y.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/d7/6d7y
https://data.pdbj.org/pub/pdb/validation_reports/d7/6d7y ftp://data.pdbj.org/pub/pdb/validation_reports/d7/6d7y
ftp://data.pdbj.org/pub/pdb/validation_reports/d7/6d7y Links
Links Assembly
Assembly


 Components
Components

 Enterobacter cloacae (bacteria) / Gene: SAMEA2273162_06031 / Plasmid: pMCSG53 / Production host:
Enterobacter cloacae (bacteria) / Gene: SAMEA2273162_06031 / Plasmid: pMCSG53 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 21-ID-F / Wavelength: 0.97872 Å
/ Beamline: 21-ID-F / Wavelength: 0.97872 Å Processing
Processing SAD / Resolution: 1.75→29.18 Å / Cor.coef. Fo:Fc: 0.965 / Cor.coef. Fo:Fc free: 0.95 / SU B: 7.205 / SU ML: 0.109 / Cross valid method: THROUGHOUT / ESU R: 0.141 / ESU R Free: 0.13 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
SAD / Resolution: 1.75→29.18 Å / Cor.coef. Fo:Fc: 0.965 / Cor.coef. Fo:Fc free: 0.95 / SU B: 7.205 / SU ML: 0.109 / Cross valid method: THROUGHOUT / ESU R: 0.141 / ESU R Free: 0.13 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller









 PDBj
PDBj