+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6c3s | ||||||
|---|---|---|---|---|---|---|---|
| Title | AMYLOID FORMING PEPTIDE YTIAAL FROM TRANSTHYRETIN | ||||||
 Components Components | TYR-THR-ILE-ALA-ALA-LEU | ||||||
 Keywords Keywords | PROTEIN FIBRIL / amyloid / transthyretin / fibril | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.602 Å MOLECULAR REPLACEMENT / Resolution: 1.602 Å | ||||||
 Authors Authors | Sievers, S.A. / Sawaya, M.R. / Saelices, L. / Eisenberg, D.S. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2018 Journal: Protein Sci. / Year: 2018Title: Crystal structures of amyloidogenic segments of human transthyretin. Authors: Saelices, L. / Sievers, S.A. / Sawaya, M.R. / Eisenberg, D.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6c3s.cif.gz 6c3s.cif.gz | 9.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6c3s.ent.gz pdb6c3s.ent.gz | 4.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6c3s.json.gz 6c3s.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c3/6c3s https://data.pdbj.org/pub/pdb/validation_reports/c3/6c3s ftp://data.pdbj.org/pub/pdb/validation_reports/c3/6c3s ftp://data.pdbj.org/pub/pdb/validation_reports/c3/6c3s | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 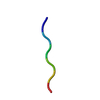
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | x 20
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein/peptide | Mass: 650.764 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.54 Å3/Da / Density % sol: 20.38 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 5.5 Details: YTIAAL crystals were grown from a solution containing 10mg/mL peptide. The reservoir contained 100mM Bis-Tris pH 5.5 and 3 M sodium chloride. Crystals were soaked on 25% Glycerol, prior to diffraction |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID13 / Wavelength: 0.895432 Å / Beamline: ID13 / Wavelength: 0.895432 Å | ||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Jul 13, 2007 | ||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.895432 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.6→90 Å / Num. obs: 625 / % possible obs: 98.9 % / Redundancy: 5.6 % / Biso Wilson estimate: 10.22 Å2 / Rmerge(I) obs: 0.103 / Χ2: 1.006 / Net I/σ(I): 9.6 / Num. measured all: 3501 | ||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: Ideal beta strand Resolution: 1.602→22.398 Å / SU ML: 0.08 / Cross valid method: THROUGHOUT / σ(F): 1.4 / Phase error: 28.03
| ||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||
| Displacement parameters | Biso max: 28.35 Å2 / Biso mean: 13.43 Å2 / Biso min: 4.56 Å2 | ||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.602→22.398 Å
| ||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.6022→1.659 Å / Rfactor Rfree error: 0 / Total num. of bins used: 1
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj

