Entry Database : PDB / ID : 6aqfTitle Crystal structure of A2AAR-BRIL in complex with the antagonist ZM241385 produced from Pichia pastoris Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a Keywords / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Escherichia coli (E. coli)Method / / / Resolution : 2.51 Å Authors Eddy, M.T. / Lee, M.Y. / Gao, Z. / White, K. / Didenko, T. / Horst, R. / Audet, M. / Stanczak, P. / McClary, K.M. / Han, G.W. ...Eddy, M.T. / Lee, M.Y. / Gao, Z. / White, K. / Didenko, T. / Horst, R. / Audet, M. / Stanczak, P. / McClary, K.M. / Han, G.W. / Jacobson, K.A. / Stevens, R.C. / Wuthrich, K. Journal : Cell / Year : 2018Title : Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor.Authors : Eddy, M.T. / Lee, M.Y. / Gao, Z.G. / White, K.L. / Didenko, T. / Horst, R. / Audet, M. / Stanczak, P. / McClary, K.M. / Han, G.W. / Jacobson, K.A. / Stevens, R.C. / Wuthrich, K. History Deposition Aug 19, 2017 Deposition site / Processing site Revision 1.0 Jan 10, 2018 Provider / Type Revision 1.1 Jan 17, 2018 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.name Revision 1.2 Jan 24, 2018 Group / Category Item _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.year Revision 1.3 Oct 4, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accessionRevision 1.4 Oct 16, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human)
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.51 Å
MOLECULAR REPLACEMENT / Resolution: 2.51 Å  Authors
Authors Citation
Citation Journal: Cell / Year: 2018
Journal: Cell / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6aqf.cif.gz
6aqf.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6aqf.ent.gz
pdb6aqf.ent.gz PDB format
PDB format 6aqf.json.gz
6aqf.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/aq/6aqf
https://data.pdbj.org/pub/pdb/validation_reports/aq/6aqf ftp://data.pdbj.org/pub/pdb/validation_reports/aq/6aqf
ftp://data.pdbj.org/pub/pdb/validation_reports/aq/6aqf
 Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human), (gene. exp.)
Homo sapiens (human), (gene. exp.) 
 Pichia (fungus) / References: UniProt: P29274, UniProt: P0ABE7
Pichia (fungus) / References: UniProt: P29274, UniProt: P0ABE7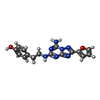









 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 23-ID-D / Wavelength: 1.033 Å
/ Beamline: 23-ID-D / Wavelength: 1.033 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj





















