[English] 日本語
 Yorodumi
Yorodumi- PDB-6aee: Crystal structure of the four Ig-like domains of LILRB1 complexed... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6aee | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | ||||||
 Components Components |
| ||||||
 Keywords Keywords | IMMUNE SYSTEM / leukocyte immunoglobulin-like receptor / immunotherapy | ||||||
| Function / homology |  Function and homology information Function and homology informationperipheral B cell tolerance induction / positive regulation of tolerance induction / HLA-A specific inhibitory MHC class I receptor activity / positive regulation of gamma-delta T cell activation involved in immune response / MHC class Ib protein complex binding / inhibitory MHC class I receptor activity / HLA-B specific inhibitory MHC class I receptor activity / immune response-inhibiting cell surface receptor signaling pathway / negative regulation of dendritic cell differentiation / Fc receptor mediated inhibitory signaling pathway ...peripheral B cell tolerance induction / positive regulation of tolerance induction / HLA-A specific inhibitory MHC class I receptor activity / positive regulation of gamma-delta T cell activation involved in immune response / MHC class Ib protein complex binding / inhibitory MHC class I receptor activity / HLA-B specific inhibitory MHC class I receptor activity / immune response-inhibiting cell surface receptor signaling pathway / negative regulation of dendritic cell differentiation / Fc receptor mediated inhibitory signaling pathway / MHC class Ib protein binding / MHC class Ib receptor activity / positive regulation of natural killer cell cytokine production / negative regulation of T cell mediated cytotoxicity / immune response-regulating signaling pathway / negative regulation of CD8-positive, alpha-beta T cell activation / MHC class I receptor activity / cis-Golgi network membrane / negative regulation of transforming growth factor beta production / interleukin-10-mediated signaling pathway / negative regulation of immune response / negative regulation of cytokine production involved in immune response / negative regulation of alpha-beta T cell activation / positive regulation of T cell tolerance induction / negative regulation of serotonin secretion / dendritic cell differentiation / protein phosphatase 1 binding / negative regulation of osteoclast development / negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell / negative regulation of G0 to G1 transition / negative regulation of interleukin-12 production / negative regulation of dendritic cell apoptotic process / negative regulation of endocytosis / negative regulation of interferon-beta production / negative regulation of mononuclear cell proliferation / negative regulation of natural killer cell mediated cytotoxicity / filopodium membrane / positive regulation of regulatory T cell differentiation / positive regulation of macrophage cytokine production / CD8 receptor binding / negative regulation of interleukin-10 production / protein homotrimerization / MHC class I protein binding / negative regulation of calcium ion transport / negative regulation of type II interferon production / T cell proliferation involved in immune response / negative regulation of cell cycle / protection from natural killer cell mediated cytotoxicity / negative regulation of tumor necrosis factor production / positive regulation of endothelial cell apoptotic process / antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent / antigen processing and presentation of endogenous peptide antigen via MHC class Ib / cellular defense response / protein localization to CENP-A containing chromatin / CENP-A containing nucleosome / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / negative regulation of T cell proliferation / positive regulation of defense response to virus by host / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / Deposition of new CENPA-containing nucleosomes at the centromere / positive regulation of interleukin-12 production / Inhibition of DNA recombination at telomere / Meiotic synapsis / RNA Polymerase I Promoter Opening / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / Assembly of the ORC complex at the origin of replication / SH2 domain binding / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / DNA methylation / : / : / Condensation of Prophase Chromosomes / negative regulation of angiogenesis / positive regulation of receptor binding / Chromatin modifications during the maternal to zygotic transition (MZT) / early endosome lumen / HCMV Late Events / SIRT1 negatively regulates rRNA expression / Nef mediated downregulation of MHC class I complex cell surface expression / negative regulation of receptor binding / DAP12 interactions / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / cellular response to iron ion / lumenal side of endoplasmic reticulum membrane / Endosomal/Vacuolar pathway / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / HDACs deacetylate histones / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / RNA Polymerase I Promoter Escape / peptide antigen assembly with MHC class II protein complex / Transcriptional regulation by small RNAs / cellular response to iron(III) ion / antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent / MHC class II protein complex Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.303 Å MOLECULAR REPLACEMENT / Resolution: 3.303 Å | ||||||
 Authors Authors | Wang, Q. / Song, H. / Qi, J. / Gao, G.F. | ||||||
 Citation Citation |  Journal: Cell. Mol. Immunol. / Year: 2019 Journal: Cell. Mol. Immunol. / Year: 2019Title: Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex. Authors: Wang, Q. / Song, H. / Cheng, H. / Qi, J. / Nam, G. / Tan, S. / Wang, J. / Fang, M. / Shi, Y. / Tian, Z. / Cao, X. / An, Z. / Yan, J. / Gao, G.F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6aee.cif.gz 6aee.cif.gz | 305.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6aee.ent.gz pdb6aee.ent.gz | 247.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6aee.json.gz 6aee.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ae/6aee https://data.pdbj.org/pub/pdb/validation_reports/ae/6aee ftp://data.pdbj.org/pub/pdb/validation_reports/ae/6aee ftp://data.pdbj.org/pub/pdb/validation_reports/ae/6aee | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6aedC  2dypS 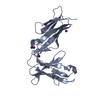 4ll9S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 32046.559 Da / Num. of mol.: 2 / Fragment: UNP residues 25-300 / Mutation: C42S, L110I, Q115R Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: HLA-G / Production host: Homo sapiens (human) / Gene: HLA-G / Production host:  #2: Protein | Mass: 11879.356 Da / Num. of mol.: 2 / Fragment: UNP residues 21-119 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: B2M / Production host: Homo sapiens (human) / Gene: B2M / Production host:  #3: Protein/peptide | Mass: 1148.424 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) / References: UniProt: P04908*PLUS Homo sapiens (human) / References: UniProt: P04908*PLUS#4: Protein | Mass: 44143.148 Da / Num. of mol.: 2 / Fragment: UNP residues 25-417 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: LILRB1 / Production host: Homo sapiens (human) / Gene: LILRB1 / Production host:  Trichoplusia ni (cabbage looper) / Variant (production host): Hi5 / References: UniProt: A0A0G2JQ44, UniProt: Q8NHL6*PLUS Trichoplusia ni (cabbage looper) / Variant (production host): Hi5 / References: UniProt: A0A0G2JQ44, UniProt: Q8NHL6*PLUSHas protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.9 Å3/Da / Density % sol: 57.56 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / Details: 0.2M imidazole malate, 15%(w/v) PEG4000, pH 6.0 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL17U1 / Wavelength: 0.97922 Å / Beamline: BL17U1 / Wavelength: 0.97922 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Oct 10, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97922 Å / Relative weight: 1 |
| Reflection | Resolution: 3.3→50 Å / Num. obs: 29866 / % possible obs: 99.9 % / Redundancy: 4.2 % / CC1/2: 0.972 / Rpim(I) all: 0.105 / Net I/σ(I): 7.89 |
| Reflection shell | Resolution: 3.3→3.42 Å / Num. unique obs: 2993 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2DYP, 4LL9 Resolution: 3.303→45.42 Å / SU ML: 0.28 / Cross valid method: FREE R-VALUE / σ(F): 1.37 / Phase error: 27.16 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.303→45.42 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj













