[English] 日本語
 Yorodumi
Yorodumi- PDB-5zmc: Structural Basis for Reactivation of -146C>T Mutant TERT Promoter... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5zmc | ||||||
|---|---|---|---|---|---|---|---|
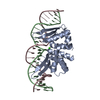
| Title | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION/DNA / ETS1 / p52 / transcription factor / TRANSCRIPTION / -146c>T mutant TERT promoter activation / TRANSCRIPTION-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationfollicular dendritic cell differentiation / Bcl3/NF-kappaB2 complex / DEx/H-box helicases activate type I IFN and inflammatory cytokines production / IkBA variant leads to EDA-ID / PML body organization / SUMOylation of immune response proteins / RIP-mediated NFkB activation via ZBP1 / Interleukin-1 processing / germinal center formation / positive regulation of leukocyte adhesion to vascular endothelial cell ...follicular dendritic cell differentiation / Bcl3/NF-kappaB2 complex / DEx/H-box helicases activate type I IFN and inflammatory cytokines production / IkBA variant leads to EDA-ID / PML body organization / SUMOylation of immune response proteins / RIP-mediated NFkB activation via ZBP1 / Interleukin-1 processing / germinal center formation / positive regulation of leukocyte adhesion to vascular endothelial cell / non-canonical NF-kappaB signal transduction / negative regulation of cell cycle / TRAF6 mediated NF-kB activation / positive regulation of blood vessel endothelial cell migration / The NLRP3 inflammasome / regulation of angiogenesis / canonical NF-kappaB signal transduction / Purinergic signaling in leishmaniasis infection / spleen development / response to cytokine / extracellular matrix organization / positive regulation of endothelial cell migration / positive regulation of erythrocyte differentiation / transcription corepressor binding / NIK-->noncanonical NF-kB signaling / cell motility / Dectin-1 mediated noncanonical NF-kB signaling / TAK1-dependent IKK and NF-kappa-B activation / Oncogene Induced Senescence / positive regulation of miRNA transcription / PKMTs methylate histone lysines / positive regulation of angiogenesis / positive regulation of inflammatory response / sequence-specific double-stranded DNA binding / rhythmic process / DNA-binding transcription activator activity, RNA polymerase II-specific / regulation of apoptotic process / response to lipopolysaccharide / nucleic acid binding / transcription by RNA polymerase II / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / cell differentiation / transcription cis-regulatory region binding / immune response / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of cell population proliferation / response to antibiotic / positive regulation of gene expression / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / chromatin / positive regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.99 Å MOLECULAR REPLACEMENT / Resolution: 2.99 Å | ||||||
 Authors Authors | Xu, X. / Bharath, S.R. / Song, H. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1. Authors: Xu, X. / Li, Y. / Bharath, S.R. / Ozturk, M.B. / Bowler, M.W. / Loo, B.Z.L. / Tergaonkar, V. / Song, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5zmc.cif.gz 5zmc.cif.gz | 143.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5zmc.ent.gz pdb5zmc.ent.gz | 108.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5zmc.json.gz 5zmc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zm/5zmc https://data.pdbj.org/pub/pdb/validation_reports/zm/5zmc ftp://data.pdbj.org/pub/pdb/validation_reports/zm/5zmc ftp://data.pdbj.org/pub/pdb/validation_reports/zm/5zmc | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 5014.243 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) |
|---|---|
| #2: DNA chain | Mass: 4787.080 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human) |
| #3: Protein | Mass: 13114.947 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ETS1, EWSR2 / Production host: Homo sapiens (human) / Gene: ETS1, EWSR2 / Production host:  |
| #4: Protein | Mass: 33165.973 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: NFKB2, LYT10 / Production host: Homo sapiens (human) / Gene: NFKB2, LYT10 / Production host:  |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.01 Å3/Da / Density % sol: 59.1 % |
|---|---|
| Crystal grow | Temperature: 290 K / Method: vapor diffusion, hanging drop / Details: 0.1 M HEPES pH 7.0 and 2.0 M Ammonium sulphate |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: MASSIF-1 / Wavelength: 1 Å / Beamline: MASSIF-1 / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS3 2M / Detector: PIXEL / Date: Jan 27, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.99→71.41 Å / Num. obs: 15992 / % possible obs: 99.9 % / Redundancy: 6.7 % / CC1/2: 0.99 / Rmerge(I) obs: 0.06 / Net I/σ(I): 20.3 |
| Reflection shell | Resolution: 2.99→3.22 Å / Redundancy: 7.2 % / Rmerge(I) obs: 0.8 / Num. unique obs: 2514 / CC1/2: 0.82 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1A3Q, 1K78 Resolution: 2.99→71.41 Å / Cor.coef. Fo:Fc: 0.902 / Cor.coef. Fo:Fc free: 0.875 / SU B: 38.236 / SU ML: 0.316 / Cross valid method: THROUGHOUT / ESU R: 0.755 / ESU R Free: 0.4 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.9 Å / Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 101.287 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2.99→71.41 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj



















































