Entry Database : PDB / ID : 5zhwTitle VanYB in complex with D-Alanine-D-Alanine D-alanyl-D-alanine carboxypeptidase Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / Biological species Enterococcus faecalis V583 (bacteria)Method / / / Resolution : 2.18 Å Authors Kim, H.S. / Hahn, H. Funding support Organization Grant number Country Korea Ministry of Science, ICT and Future Planning 2013R1A2A1A05067303 Korea Ministry of Science and ICT NRF-2011-0030001 Korea Ministry of Science and ICT NRF-2013M3A6A4043695 Korea Ministry of Science and ICT NRF-2017R1C1B2012225 National Cancer Center Grant of Korea NCC-1120170 Korea Ministry of Education NRF-2017R1A6A3A01002416
Journal : Int. J. Biol. Macromol. / Year : 2018Title : Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.Authors : Kim, H.S. / Hahn, H. / Kim, J. / Jang, D.M. / Lee, J.Y. / Back, J.M. / Im, H.N. / Kim, H. / Han, B.W. / Suh, S.W. History Deposition Mar 13, 2018 Deposition site / Processing site Revision 1.0 Sep 5, 2018 Provider / Type Revision 1.1 Nov 22, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id Revision 1.2 Oct 23, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Enterococcus faecalis V583 (bacteria)
Enterococcus faecalis V583 (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.18 Å
MOLECULAR REPLACEMENT / Resolution: 2.18 Å  Authors
Authors Korea, Republic Of, 6items
Korea, Republic Of, 6items  Citation
Citation Journal: Int. J. Biol. Macromol. / Year: 2018
Journal: Int. J. Biol. Macromol. / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5zhw.cif.gz
5zhw.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5zhw.ent.gz
pdb5zhw.ent.gz PDB format
PDB format 5zhw.json.gz
5zhw.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/zh/5zhw
https://data.pdbj.org/pub/pdb/validation_reports/zh/5zhw ftp://data.pdbj.org/pub/pdb/validation_reports/zh/5zhw
ftp://data.pdbj.org/pub/pdb/validation_reports/zh/5zhw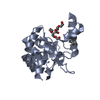

 Links
Links Assembly
Assembly


 Components
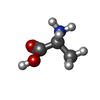
Components Enterococcus faecalis V583 (bacteria) / Strain: ATCC 700802 / V583 / Gene: vanYB, EF_2297 / Production host:
Enterococcus faecalis V583 (bacteria) / Strain: ATCC 700802 / V583 / Gene: vanYB, EF_2297 / Production host: 








 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Photon Factory
Photon Factory  / Beamline: BL-1A / Wavelength: 1 Å
/ Beamline: BL-1A / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj



