+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5yip | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of AnkG LIR/GABARAPL1 complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / Autophagy | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of protein targeting / positive regulation of cell communication by electrical coupling / positive regulation of membrane depolarization during cardiac muscle cell action potential / maintenance of protein location in plasma membrane / positive regulation of sodium ion import across plasma membrane / substrate localization to autophagosome / membrane assembly / Macroautophagy / protein localization to axon / clustering of voltage-gated sodium channels ...regulation of protein targeting / positive regulation of cell communication by electrical coupling / positive regulation of membrane depolarization during cardiac muscle cell action potential / maintenance of protein location in plasma membrane / positive regulation of sodium ion import across plasma membrane / substrate localization to autophagosome / membrane assembly / Macroautophagy / protein localization to axon / clustering of voltage-gated sodium channels / spectrin-associated cytoskeleton / establishment or maintenance of microtubule cytoskeleton polarity / magnesium ion homeostasis / regulation of potassium ion transport / positive regulation of membrane potential / glycophagy / plasma membrane organization / negative regulation of delayed rectifier potassium channel activity / channel activator activity / maintenance of protein location in cell / phosphorylation-dependent protein binding / positive regulation of homotypic cell-cell adhesion / paranode region of axon / positive regulation of sodium ion transport / regulation of modification of postsynaptic structure / negative regulation of endocytosis / costamere / axon initial segment / anterograde axonal transport / GABA receptor binding / cellular response to magnesium ion / Tat protein binding / phosphatidylethanolamine binding / Golgi to plasma membrane protein transport / node of Ranvier / cellular response to nitrogen starvation / spectrin binding / axon development / neuromuscular junction development / response to immobilization stress / mitotic cytokinesis / autophagosome membrane / intercalated disc / lateral plasma membrane / sodium channel regulator activity / autophagosome maturation / autophagosome assembly / positive regulation of protein targeting to membrane / bicellular tight junction / neuronal action potential / mitophagy / cytoskeletal protein binding / protein-membrane adaptor activity / axon cytoplasm / T-tubule / dendrite membrane / dendrite cytoplasm / axonogenesis / autophagosome / axon guidance / basal plasma membrane / cytoplasmic vesicle membrane / sarcoplasmic reticulum / protein localization to plasma membrane / neuromuscular junction / establishment of protein localization / positive regulation of non-canonical NF-kappaB signal transduction / sarcolemma / synapse organization / phospholipid binding / structural constituent of cytoskeleton / Z disc / cell body / basolateral plasma membrane / protein-macromolecule adaptor activity / microtubule / RNA polymerase II-specific DNA-binding transcription factor binding / transmembrane transporter binding / cytoskeleton / postsynaptic membrane / lysosome / neuron projection / postsynapse / postsynaptic density / cilium / ciliary basal body / cadherin binding / axon / ubiquitin protein ligase binding / dendrite / synapse / positive regulation of gene expression / regulation of transcription by RNA polymerase II / glutamatergic synapse / cell surface / endoplasmic reticulum / Golgi apparatus / signal transduction / mitochondrion / membrane Similarity search - Function | ||||||
| Biological species |   | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.85 Å MOLECULAR REPLACEMENT / Resolution: 1.85 Å | ||||||
 Authors Authors | Li, J. / Zhu, R. / Chen, K. / Zheng, H. / Yuan, C. / Zhang, H. / Wang, C. / Zhang, M. | ||||||
| Funding support |  Hong Kong, 1items Hong Kong, 1items
| ||||||
 Citation Citation |  Journal: Nat. Chem. Biol. / Year: 2018 Journal: Nat. Chem. Biol. / Year: 2018Title: Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Authors: Li, J. / Zhu, R. / Chen, K. / Zheng, H. / Zhao, H. / Yuan, C. / Zhang, H. / Wang, C. / Zhang, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5yip.cif.gz 5yip.cif.gz | 77.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5yip.ent.gz pdb5yip.ent.gz | 56 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5yip.json.gz 5yip.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yi/5yip https://data.pdbj.org/pub/pdb/validation_reports/yi/5yip ftp://data.pdbj.org/pub/pdb/validation_reports/yi/5yip ftp://data.pdbj.org/pub/pdb/validation_reports/yi/5yip | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5yiqC  5yirC  5yisC  2r2qS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 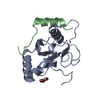
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 14642.653 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein/peptide | Mass: 2919.998 Da / Num. of mol.: 1 / Fragment: UNP RESIDUES 1985-2010 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #3: Chemical | ChemComp-GOL / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.85 Å3/Da / Density % sol: 56.83 % |
|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion / pH: 4.6 Details: 35% w/v Pentaerythritol ethoxylate 797 (15/4 EO/OH), 0.2 M ammonium sulfate, 0.1 M sodium acetate (pH 4.6) |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL19U1 / Wavelength: 0.97774 Å / Beamline: BL19U1 / Wavelength: 0.97774 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Oct 20, 2015 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97774 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.85→50 Å / Num. obs: 18077 / % possible obs: 100 % / Redundancy: 15.9 % / Biso Wilson estimate: 27.51 Å2 / Rmerge(I) obs: 0.084 / Rpim(I) all: 0.022 / Rrim(I) all: 0.087 / Χ2: 0.529 / Net I/σ(I): 4.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2R2Q Resolution: 1.85→30.118 Å / SU ML: 0.21 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 20.6
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 110.07 Å2 / Biso mean: 37.7709 Å2 / Biso min: 17.03 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.85→30.118 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 6 / % reflection obs: 100 %
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj










