[English] 日本語
 Yorodumi
Yorodumi- PDB-5wbf: Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from H... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5wbf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from Helicobacter pylori | |||||||||
 Components Components | Methyl-accepting chemotaxis transducer (TlpC) | |||||||||
 Keywords Keywords | SIGNALING PROTEIN / Bacterial protein / Chemoreceptor sensing domain / double-CACHE domain / Helicobacter pylori | |||||||||
| Function / homology | Methyl-accepting chemotaxis protein (MCP) signalling domain / Methyl-accepting chemotaxis protein (MCP) signalling domain / Bacterial chemotaxis sensory transducers domain profile. / Methyl-accepting chemotaxis-like domains (chemotaxis sensory transducer). / chemotaxis / signal transduction / membrane / LACTIC ACID / Methyl-accepting chemotaxis transducer (TlpC) Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / SAD /  molecular replacement / Resolution: 2.19 Å molecular replacement / Resolution: 2.19 Å | |||||||||
 Authors Authors | Machuca, M.A. / Johnson, K.S. / Liu, Y.C. / Steer, D.L. / Ottemann, K.M. / Roujeinikova, A. | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2017 Journal: Sci Rep / Year: 2017Title: Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate. Authors: Machuca, M.A. / Johnson, K.S. / Liu, Y.C. / Steer, D.L. / Ottemann, K.M. / Roujeinikova, A. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5wbf.cif.gz 5wbf.cif.gz | 178.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5wbf.ent.gz pdb5wbf.ent.gz | 141.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5wbf.json.gz 5wbf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wb/5wbf https://data.pdbj.org/pub/pdb/validation_reports/wb/5wbf ftp://data.pdbj.org/pub/pdb/validation_reports/wb/5wbf ftp://data.pdbj.org/pub/pdb/validation_reports/wb/5wbf | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
|
- Components
Components
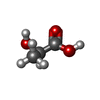
| #1: Protein | Mass: 30602.799 Da / Num. of mol.: 3 Fragment: Double CACHE (dCACHE) sensing domain (UNP residues 34-297) Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | #3: Chemical | ChemComp-GOL / | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.25 Å3/Da / Density % sol: 62.18 % |
|---|---|
| Crystal grow | Temperature: 291.15 K / Method: vapor diffusion, hanging drop Details: 0.2 M MgCl2, 0.1 M MES/NaOH (pH 6.5), 22% (w/v) PEG 4000 and 10 mM BaCl2H4O2 |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX2 / Wavelength: 1.5498 Å / Beamline: MX2 / Wavelength: 1.5498 Å | |||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Apr 10, 2014 | |||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5498 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.19→30.57 Å / Num. obs: 59028 / % possible obs: 97.6 % / Redundancy: 3.6 % / Biso Wilson estimate: 35.78 Å2 / CC1/2: 0.995 / Rmerge(I) obs: 0.062 / Rpim(I) all: 0.038 / Rrim(I) all: 0.073 / Net I/σ(I): 13.3 | |||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 2.19→30.57 Å / Cross valid method: FREE R-VALUE SAD / Resolution: 2.19→30.57 Å / Cross valid method: FREE R-VALUE
| |||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.19→30.57 Å
| |||||||||||||||||||||
| LS refinement shell | Resolution: 2.19→2.31 Å |
 Movie
Movie Controller
Controller












 PDBj
PDBj