| Entry | Database: PDB / ID: 5n4t
|
|---|
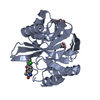
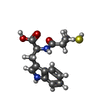
| Title | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-L-tryptophan (Compound 4) |
|---|
 Components Components | Beta-lactamase VIM-2 |
|---|
 Keywords Keywords | HYDROLASE / metallo-beta-lactamase / inhibitor / complex / antibiotic resistance |
|---|
| Function / homology |  Function and homology information Function and homology information
: / : / Metallo-beta-lactamase superfamily / Ribonuclease Z/Hydroxyacylglutathione hydrolase-like / Metallo-beta-lactamase superfamily / Metallo-beta-lactamase; Chain A / Metallo-beta-lactamase / Ribonuclease Z/Hydroxyacylglutathione hydrolase-like / 4-Layer Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.16 Å molecular replacement / Resolution: 1.16 Å |
|---|
 Authors Authors | Li, G.-B. / Brem, J. / McDonough, M.A. / Schofield, C.J. |
|---|
 Citation Citation |  Journal: Chem. Commun. (Camb.) / Year: 2017 Journal: Chem. Commun. (Camb.) / Year: 2017
Title: Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Authors: Li, G.B. / Brem, J. / Lesniak, R. / Abboud, M.I. / Lohans, C.T. / Clifton, I.J. / Yang, S.Y. / Jimenez-Castellanos, J.C. / Avison, M.B. / Spencer, J. / McDonough, M.A. / Schofield, C.J. |
|---|
| History | | Deposition | Feb 11, 2017 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | May 17, 2017 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jun 7, 2017 | Group: Database references |
|---|
| Revision 1.2 | Jan 17, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_alt_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_alt_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_ptnr2_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_symmetry |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.16 Å
molecular replacement / Resolution: 1.16 Å  Authors
Authors Citation
Citation Journal: Chem. Commun. (Camb.) / Year: 2017
Journal: Chem. Commun. (Camb.) / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5n4t.cif.gz
5n4t.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5n4t.ent.gz
pdb5n4t.ent.gz PDB format
PDB format 5n4t.json.gz
5n4t.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 5n4t_validation.pdf.gz
5n4t_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 5n4t_full_validation.pdf.gz
5n4t_full_validation.pdf.gz 5n4t_validation.xml.gz
5n4t_validation.xml.gz 5n4t_validation.cif.gz
5n4t_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/n4/5n4t
https://data.pdbj.org/pub/pdb/validation_reports/n4/5n4t ftp://data.pdbj.org/pub/pdb/validation_reports/n4/5n4t
ftp://data.pdbj.org/pub/pdb/validation_reports/n4/5n4t



 Links
Links Assembly
Assembly


 Components
Components









 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04 / Wavelength: 0.92819 Å
/ Beamline: I04 / Wavelength: 0.92819 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj




