+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5lue | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
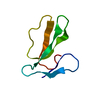
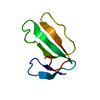
| Title | Minor form of the recombinant cytotoxin-1 from N. oxiana | |||||||||||||||||||||
 Components Components | VC-1=CYTOTOXIN | |||||||||||||||||||||
 Keywords Keywords | TOXIN / cytolytic peptide / all-beta sheet protein / cobra venom / Structure from MOLMOL | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationother organism cell membrane / toxin activity / killing of cells of another organism / extracellular region / membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Naja oxiana (Central Asian cobra) Naja oxiana (Central Asian cobra) | |||||||||||||||||||||
| Method | SOLUTION NMR / restrained Rosetta minimization | |||||||||||||||||||||
 Authors Authors | Dubovskii, P.V. / Dubinnyi, M.A. / Shulepko, M.A. / Lyukmanova, E.N. / Dolgikh, D.A. / Kirpichnikov, M.P. / Efremov, R.G. | |||||||||||||||||||||
| Funding support |  Russian Federation, 6items Russian Federation, 6items
| |||||||||||||||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2017 Journal: Biochemistry / Year: 2017Title: Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They? Authors: Dubovskii, P.V. / Dubinnyi, M.A. / Konshina, A.G. / Kazakova, E.D. / Sorokoumova, G.M. / Ilyasova, T.M. / Shulepko, M.A. / Chertkova, R.V. / Lyukmanova, E.N. / Dolgikh, D.A. / Arseniev, A.S. / Efremov, R.G. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Towards universal approach for bacterial production of three-finger Ly6/uPAR proteins: case study of Cytotoxin I from cobra N. oxiana Authors: Shulepko, M.A. / Lyukmanova, E.N. / Shenkarev, Z.O. / Dubovskii, P.V. / Astapova, M.V. / Feofanov, A.V. / Arseniev, A.S. / Utkin, Y.N. / Kirpichnikov, M.P. / Dolgikh, D.A. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5lue.cif.gz 5lue.cif.gz | 385.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5lue.ent.gz pdb5lue.ent.gz | 320.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5lue.json.gz 5lue.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lu/5lue https://data.pdbj.org/pub/pdb/validation_reports/lu/5lue ftp://data.pdbj.org/pub/pdb/validation_reports/lu/5lue ftp://data.pdbj.org/pub/pdb/validation_reports/lu/5lue | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5npnC  5t8aC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6962.535 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: The toxin gene encoding 60 amino acid residues with additional ATG codon at the 5(prime)-end was constructed from six overlapping synthetic oligonucleotides using PCR Source: (gene. exp.)  Naja oxiana (Central Asian cobra) / Production host: Naja oxiana (Central Asian cobra) / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 1 mM [U-99% 13C; U-99% 15N] recombinant cytotoxin-1, 95 v/v non-labeled H2O, 5 v/v 99.9%-2H D2O, 3 uM non-labeled NaOH, 1 uM non-labeled HCl, 95% H2O/5% D2O Details: uniformly labeled 13C-15N recombinant toxin was dissolved in H2O/D2O(95:5 v/v) mixture and pH was adjusted with small additions of concentrated NaOH, HCl solutions Label: 13C-15N sample / Solvent system: 95% H2O/5% D2O | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||
| Sample conditions | Details: identical to those used at the investigation of the native toxin (deposited under code of 1RL5) Ionic strength: 0 Not defined / Ionic strength err: 0.1 / Label: conditions_1 / pH: 6.5 / PH err: 0.1 / Pressure: 101325 Pa / Pressure err: 100 / Temperature: 303 K / Temperature err: 0.2 |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: restrained Rosetta minimization / Software ordinal: 4 Details: as in: J Am Chem Soc. 2014 Feb 5;136(5):1893-906. Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures. Mao B, Tejero R, Baker D, Montelione GT. | ||||||||||||||||||||||||
| NMR representative | Selection criteria: quality score | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 200 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj

 HNCA
HNCA