[English] 日本語
 Yorodumi
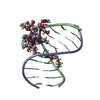
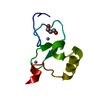
Yorodumi- PDB-2hp8: SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENC... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2hp8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, 30 STRUCTURES | |||||||||
 Components Components | Cx9C motif-containing protein 4 | |||||||||
 Keywords Keywords | CYSTEINE MOTIF / HU-P8 / LEUKEMIA | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial protein import / mitochondrial intermembrane space / mitochondrion Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY, RESTRAINED MOLECULAR DYNAMICS | |||||||||
 Authors Authors | Barthe, P. / Chiche, L. / Strub, M.P. / Roumestand, C. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1997 Journal: J.Mol.Biol. / Year: 1997Title: Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif. Authors: Barthe, P. / Yang, Y.S. / Chiche, L. / Hoh, F. / Strub, M.P. / Guignard, L. / Soulier, J. / Stern, M.H. / van Tilbeurgh, H. / Lhoste, J.M. / Roumestand, C. #1:  Journal: Oncogene / Year: 1994 Journal: Oncogene / Year: 1994Title: The Mtcp-1/C6.1B Gene Encodes for a Cytoplasmic 8 Kd Protein Overexpressed in T Cell Leukemia Bearing a T(X;14) Translocation Authors: Soulier, J. / Madani, A. / Cacheux, V. / Rosenzwajg, M. / Sigaux, F. / Stern, M.H. #2:  Journal: Oncogene / Year: 1993 Journal: Oncogene / Year: 1993Title: Mtcp-1: A Novel Gene on the Human Chromosome Xq28 Translocated to the T Cell Receptor Alpha/Delta Locus in Mature T Cell Proliferations Authors: Stern, M.H. / Soulier, J. / Rosenzwajg, M. / Nakahara, K. / Canki-Klain, N. / Aurias, A. / Sigaux, F. / Kirsch, I.R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2hp8.cif.gz 2hp8.cif.gz | 607.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2hp8.ent.gz pdb2hp8.ent.gz | 509.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2hp8.json.gz 2hp8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hp/2hp8 https://data.pdbj.org/pub/pdb/validation_reports/hp/2hp8 ftp://data.pdbj.org/pub/pdb/validation_reports/hp/2hp8 ftp://data.pdbj.org/pub/pdb/validation_reports/hp/2hp8 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 8889.188 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Cell: T-LYMPHOCYTE / Cellular location: MITOCHONDRIA / Gene: CMC4, C6.1B, MTCP1, MTCP1NB / Plasmid: PGEX2T / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Cell: T-LYMPHOCYTE / Cellular location: MITOCHONDRIA / Gene: CMC4, C6.1B, MTCP1, MTCP1NB / Plasmid: PGEX2T / Species (production host): Escherichia coli / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 10mM PO4 mM / Label: sample conditions / pH: 6.5 / Pressure: 1 atm / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AMX / Manufacturer: Bruker / Model: AMX / Field strength: 600 MHz |
|---|
- Processing
Processing
| Software | Name:  AMBER / Classification: refinement AMBER / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||||||||||||||
| Refinement | Method: DISTANCE GEOMETRY, RESTRAINED MOLECULAR DYNAMICS / Software ordinal: 1 Details: THE 3D STRUCTURE OF P8-MTCP1 IN SOLUTION BY NMR IS BASED ON 931 APPROXIMATE INTERPROTON DISTANCE RESTRAINTS AND 57 TORSION ANGLE RESTRAINTS DERIVED FROM NOE AND COUPLING CONSTANT ...Details: THE 3D STRUCTURE OF P8-MTCP1 IN SOLUTION BY NMR IS BASED ON 931 APPROXIMATE INTERPROTON DISTANCE RESTRAINTS AND 57 TORSION ANGLE RESTRAINTS DERIVED FROM NOE AND COUPLING CONSTANT MEASUREMENTS. THE STRUCTURES ARE CALCULATED USING THE DISTANCE GEOMETRY DIANA METHOD AND REFINED USING A SIMULATED-ANNEALING PROTOCOL WITH THE PROGRAM AMBER 4.1. | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 30 / Conformers submitted total number: 30 |
 Movie
Movie Controller
Controller










 PDBj
PDBj

