Entry Database : PDB / ID : 5kohTitle Nitrogenase MoFeP from Gluconacetobacter diazotrophicus in dithionite reduced state Nitrogenase FeMo beta subunit protein NifK Nitrogenase protein alpha chain Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Gluconacetobacter diazotrophicus (bacteria)Method / / / Resolution : 1.83 Å Authors Owens, C.P. / Tezcan, F.A. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM099813 Frasch Foundation 735-HF12 National Institute of Food and Agriculture (NIFA, United States) 2015-67012-22895
Journal : J.Am.Chem.Soc. / Year : 2016Title : Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer.Authors : Owens, C.P. / Katz, F.E. / Carter, C.H. / Oswald, V.F. / Tezcan, F.A. History Deposition Jun 30, 2016 Deposition site / Processing site Revision 1.0 Sep 21, 2016 Provider / Type Revision 1.1 Aug 23, 2017 Group / Derived calculations / Category / pdbx_struct_oper_listItem / _pdbx_struct_oper_list.symmetry_operationRevision 1.2 Sep 20, 2017 Group / Refinement description / Category / softwareItem / _software.classificationRevision 1.3 Dec 25, 2019 Group / Derived calculations / Category / pdbx_struct_special_symmetry / Item Revision 1.4 Mar 16, 2022 Group / Database references / Derived calculationsCategory database_2 / pdbx_audit_support ... database_2 / pdbx_audit_support / pdbx_struct_conn_angle / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_audit_support.funding_organization / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id Revision 1.5 Oct 4, 2023 Group / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / pdbx_initial_refinement_model / struct_ncs_dom_lim Item _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id ... _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Gluconacetobacter diazotrophicus (bacteria)
Gluconacetobacter diazotrophicus (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.83 Å
MOLECULAR REPLACEMENT / Resolution: 1.83 Å  Authors
Authors United States, 3items
United States, 3items  Citation
Citation Journal: J.Am.Chem.Soc. / Year: 2016
Journal: J.Am.Chem.Soc. / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5koh.cif.gz
5koh.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5koh.ent.gz
pdb5koh.ent.gz PDB format
PDB format 5koh.json.gz
5koh.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ko/5koh
https://data.pdbj.org/pub/pdb/validation_reports/ko/5koh ftp://data.pdbj.org/pub/pdb/validation_reports/ko/5koh
ftp://data.pdbj.org/pub/pdb/validation_reports/ko/5koh

 Links
Links Assembly
Assembly
 Components
Components Gluconacetobacter diazotrophicus (strain ATCC 49037 / DSM 5601 / PAl5) (bacteria)
Gluconacetobacter diazotrophicus (strain ATCC 49037 / DSM 5601 / PAl5) (bacteria) Gluconacetobacter diazotrophicus (strain ATCC 49037 / DSM 5601 / PAl5) (bacteria)
Gluconacetobacter diazotrophicus (strain ATCC 49037 / DSM 5601 / PAl5) (bacteria)
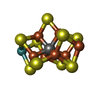





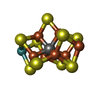





 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRL
SSRL  / Beamline: BL9-2 / Wavelength: 0.98 Å
/ Beamline: BL9-2 / Wavelength: 0.98 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller


















 PDBj
PDBj





