Entry Database : PDB / ID : 4wnaTitle Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon (Nitrogenase molybdenum-iron protein ...) x 2 Keywords / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Azotobacter vinelandii (bacteria)Method / Resolution : 2 Å Authors Morrison, C.N. / Hoy, J.A. / Zhang, L. / Einsle, O. / Rees, D.C. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM45162 Howard Hughes Medical Institute (HHMI) European Research Council (ERC) 310656 European Union German Research Foundation (DFG) Ei-520/7 German Research Foundation (DFG) IRTG 1478
Journal : Biochemistry / Year : 2015Title : Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.Authors : Morrison, C.N. / Hoy, J.A. / Zhang, L. / Einsle, O. / Rees, D.C. History Deposition Oct 11, 2014 Deposition site / Processing site Revision 1.0 Mar 18, 2015 Provider / Type Revision 1.1 Apr 1, 2015 Group Revision 1.2 Sep 13, 2017 Group Advisory / Author supporting evidence ... Advisory / Author supporting evidence / Database references / Derived calculations / Source and taxonomy Category citation / entity_src_gen ... citation / entity_src_gen / pdbx_audit_support / pdbx_struct_oper_list / pdbx_validate_close_contact Item _citation.journal_id_CSD / _entity_src_gen.pdbx_alt_source_flag ... _citation.journal_id_CSD / _entity_src_gen.pdbx_alt_source_flag / _pdbx_audit_support.funding_organization / _pdbx_struct_oper_list.symmetry_operation Revision 1.3 Sep 27, 2017 Group / Category / Item Revision 1.4 Nov 20, 2019 Group / Category Item / _pdbx_audit_support.funding_organizationRevision 1.5 Dec 27, 2023 Group / Database references / Derived calculationsCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_alt_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.pdbx_ptnr2_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Azotobacter vinelandii (bacteria)
Azotobacter vinelandii (bacteria) X-RAY DIFFRACTION / Resolution: 2 Å
X-RAY DIFFRACTION / Resolution: 2 Å  Authors
Authors United States, European Union,
United States, European Union,  Germany, 5items
Germany, 5items  Citation
Citation Journal: Biochemistry / Year: 2015
Journal: Biochemistry / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4wna.cif.gz
4wna.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4wna.ent.gz
pdb4wna.ent.gz PDB format
PDB format 4wna.json.gz
4wna.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/wn/4wna
https://data.pdbj.org/pub/pdb/validation_reports/wn/4wna ftp://data.pdbj.org/pub/pdb/validation_reports/wn/4wna
ftp://data.pdbj.org/pub/pdb/validation_reports/wn/4wna Links
Links Assembly
Assembly
 Components
Components Azotobacter vinelandii (bacteria) / Gene: nifD / Production host:
Azotobacter vinelandii (bacteria) / Gene: nifD / Production host:  Azotobacter vinelandii DJ (bacteria) / References: UniProt: P07328, nitrogenase
Azotobacter vinelandii DJ (bacteria) / References: UniProt: P07328, nitrogenase Azotobacter vinelandii (bacteria) / Gene: nifK / Production host:
Azotobacter vinelandii (bacteria) / Gene: nifK / Production host:  Azotobacter vinelandii DJ (bacteria) / References: UniProt: P07329, nitrogenase
Azotobacter vinelandii DJ (bacteria) / References: UniProt: P07329, nitrogenase
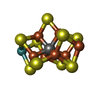




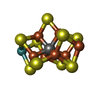




 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å Processing
Processing Movie
Movie Controller
Controller














 PDBj
PDBj





