[English] 日本語
 Yorodumi
Yorodumi- PDB-5jsh: The 3D structure of recombinant [NiFeSe] hydrogenase from Desulfo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5jsh | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The 3D structure of recombinant [NiFeSe] hydrogenase from Desulfovibrio Vulgaris Hildenborough in the oxidized state at 1.30 Angstrom | ||||||||||||
 Components Components | (Periplasmic [NiFeSe] hydrogenase, ...) x 2 | ||||||||||||
 Keywords Keywords | OXIDOREDUCTASE / hydrogenase / biological hydrogen production | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcytochrome-c3 hydrogenase / cytochrome-c3 hydrogenase activity / ferredoxin hydrogenase complex / [Ni-Fe] hydrogenase complex / ferredoxin hydrogenase activity / anaerobic respiration / 3 iron, 4 sulfur cluster binding / nickel cation binding / 4 iron, 4 sulfur cluster binding / electron transfer activity ...cytochrome-c3 hydrogenase / cytochrome-c3 hydrogenase activity / ferredoxin hydrogenase complex / [Ni-Fe] hydrogenase complex / ferredoxin hydrogenase activity / anaerobic respiration / 3 iron, 4 sulfur cluster binding / nickel cation binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / periplasmic space / metal ion binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  Desulfovibrio vulgaris (bacteria) Desulfovibrio vulgaris (bacteria) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.3 Å MOLECULAR REPLACEMENT / Resolution: 1.3 Å | ||||||||||||
 Authors Authors | Marques, M.C. / Pereira, I.A.C. / Matias, P.M. | ||||||||||||
| Funding support |  Portugal, 3items Portugal, 3items
| ||||||||||||
 Citation Citation |  Journal: Nat. Chem. Biol. / Year: 2017 Journal: Nat. Chem. Biol. / Year: 2017Title: The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis. Authors: Marques, M.C. / Tapia, C. / Gutierrez-Sanz, O. / Ramos, A.R. / Keller, K.L. / Wall, J.D. / De Lacey, A.L. / Matias, P.M. / Pereira, I.A.C. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5jsh.cif.gz 5jsh.cif.gz | 465.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5jsh.ent.gz pdb5jsh.ent.gz | 379.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5jsh.json.gz 5jsh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/js/5jsh https://data.pdbj.org/pub/pdb/validation_reports/js/5jsh ftp://data.pdbj.org/pub/pdb/validation_reports/js/5jsh ftp://data.pdbj.org/pub/pdb/validation_reports/js/5jsh | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5jskC  5jsuC  5jsyC  5jt1C  2wpnS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Periplasmic [NiFeSe] hydrogenase, ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 33940.879 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / NCIMB 8303) (bacteria) Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / NCIMB 8303) (bacteria)Gene: hysB, DVU_1917 Production host:  Desulfovibrio vulgaris str. Hildenborough (bacteria) Desulfovibrio vulgaris str. Hildenborough (bacteria)References: UniProt: Q72AS4, ferredoxin hydrogenase |
|---|---|
| #2: Protein | Mass: 55989.047 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Cysteine 75 is oxidized to sulfenate (CSD) in the crystal structure Source: (gene. exp.)  Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / NCIMB 8303) (bacteria) Desulfovibrio vulgaris (strain Hildenborough / ATCC 29579 / NCIMB 8303) (bacteria)Gene: hysA, DVU_1918 Production host:  Desulfovibrio vulgaris str. Hildenborough (bacteria) Desulfovibrio vulgaris str. Hildenborough (bacteria)References: UniProt: Q72AS3, ferredoxin hydrogenase |
-Non-polymers , 8 types, 865 molecules 
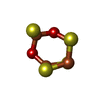



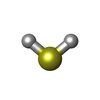






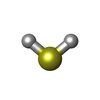


| #3: Chemical | | #4: Chemical | ChemComp-6ML / | #5: Chemical | ChemComp-FCO / | #6: Chemical | ChemComp-NI / | #7: Chemical | ChemComp-FE2 / | #8: Chemical | ChemComp-H2S / | #9: Chemical | ChemComp-CL / | #10: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.59 % / Description: irregular hexagonal plate |
|---|---|
| Crystal grow | Temperature: 295 K / Method: vapor diffusion, sitting drop / Details: 20% PEG 1500 (w/v) and 0.1 mM Tris-HCl pH 7.6 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-1 / Wavelength: 0.9334 Å / Beamline: ID14-1 / Wavelength: 0.9334 Å |
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: Nov 7, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9334 Å / Relative weight: 1 |
| Reflection | Resolution: 1.3→54 Å / Num. obs: 169426 / % possible obs: 96.9 % / Redundancy: 3.7 % / Biso Wilson estimate: 8.6 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.061 / Net I/σ(I): 13.7 |
| Reflection shell | Resolution: 1.3→1.32 Å / Redundancy: 2 % / Rmerge(I) obs: 0.315 / Mean I/σ(I) obs: 2.2 / % possible all: 70.8 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2WPN Resolution: 1.3→51.819 Å / SU ML: 0.1 / Cross valid method: FREE R-VALUE / σ(F): 0.68 / Phase error: 13.22 / Stereochemistry target values: ML Details: THE ANOMALOUS DISPERSION PARAMETERS FOR THE SE ATOM WERE REFINED. HYDROGEN ATOMS WERE ADDED IN CALCULATED POSITIONS AND REFINED IN RIDING POSITIONS. INDIVIDUAL ANISOTROPIC ATOMIC ...Details: THE ANOMALOUS DISPERSION PARAMETERS FOR THE SE ATOM WERE REFINED. HYDROGEN ATOMS WERE ADDED IN CALCULATED POSITIONS AND REFINED IN RIDING POSITIONS. INDIVIDUAL ANISOTROPIC ATOMIC DISPLACEMENT PARAMETERS FOR ALL PROTEIN NON- HYDROGEN ATOMS WERE REFINED.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.3→51.819 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj













