+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5iq5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
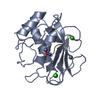
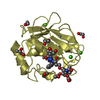
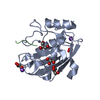
| Title | NMR solution structure of Mayaro virus macro domain | |||||||||
 Components Components | Macro domain | |||||||||
 Keywords Keywords | VIRAL PROTEIN / viral macro domains / Mayaro virus / Alphavirus / ADP ribose binding module | |||||||||
| Function / homology |  Function and homology information Function and homology informationchromatin extrusion motor activity / ATP-dependent H2AZ histone chaperone activity / ATP-dependent H3-H4 histone complex chaperone activity / cohesin loader activity / DNA clamp loader activity / ADP-ribose 1''-phosphate phosphatase / host cell filopodium / mRNA methyltransferase activity / polynucleotide 5'-phosphatase activity / mRNA 5'-phosphatase ...chromatin extrusion motor activity / ATP-dependent H2AZ histone chaperone activity / ATP-dependent H3-H4 histone complex chaperone activity / cohesin loader activity / DNA clamp loader activity / ADP-ribose 1''-phosphate phosphatase / host cell filopodium / mRNA methyltransferase activity / polynucleotide 5'-phosphatase activity / mRNA 5'-phosphatase / mRNA 5'-triphosphate monophosphatase activity / polynucleotide adenylyltransferase / poly(A) RNA polymerase activity / mRNA modification / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / 7-methylguanosine mRNA capping / cysteine-type peptidase activity / Transferases; Transferring one-carbon groups; Methyltransferases / host cell cytoplasmic vesicle membrane / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / nucleoside-triphosphate phosphatase / methylation / RNA helicase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / RNA helicase / symbiont-mediated suppression of host gene expression / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / GTP binding / host cell nucleus / host cell plasma membrane / ATP hydrolysis activity / proteolysis / RNA binding / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Mayaro virus Mayaro virus | |||||||||
| Method | SOLUTION NMR / molecular dynamics | |||||||||
 Authors Authors | Melekis, E. / Tsika, A.C. / Bentrop, D. / Papageorgiou, N. / Coutard, B. / Spyroulias, G.A. | |||||||||
| Funding support | 2items
| |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2019 Journal: J.Mol.Biol. / Year: 2019Title: Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain. Authors: Tsika, A.C. / Melekis, E. / Tsatsouli, S.A. / Papageorgiou, N. / Mate, M.J. / Canard, B. / Coutard, B. / Bentrop, D. / Spyroulias, G.A. #1: Journal: Biomol NMR Assign / Year: 2015 Title: NMR study of non-structural proteins--part I: (1)H, (13)C, (15)N backbone and side-chain resonance assignment of macro domain from Mayaro virus (MAYV). Authors: Melekis, E. / Tsika, A.C. / Lichiere, J. / Chasapis, C.T. / Margiolaki, I. / Papageorgiou, N. / Coutard, B. / Bentrop, D. / Spyroulias, G.A. #2: Journal: Biomol NMR Assign / Year: 2015 Title: NMR study of non-structural proteins--part II: (1)H, (13)C, (15)N backbone and side-chain resonance assignment of macro domain from Venezuelan equine encephalitis virus (VEEV). Authors: Makrynitsa, G.I. / Ntonti, D. / Marousis, K.D. / Tsika, A.C. / Lichiere, J. / Papageorgiou, N. / Coutard, B. / Bentrop, D. / Spyroulias, G.A. #3:  Journal: Zeitschfrift fur Kristallographie / Year: 2010 Journal: Zeitschfrift fur Kristallographie / Year: 2010Title: Preliminary insights into the non structural protein 3 macro domain of the Mayaro virus by powder diffraction Authors: Papageorgiou, N. / Watier, Y. / Saunders, L. / Coutard, B. / Lantez, V. / Gould, E.A. / Fitch, A.N. / Wright, J.P. / Canard, B. / Margiolaki, I. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5iq5.cif.gz 5iq5.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5iq5.ent.gz pdb5iq5.ent.gz | 932.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5iq5.json.gz 5iq5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/iq/5iq5 https://data.pdbj.org/pub/pdb/validation_reports/iq/5iq5 ftp://data.pdbj.org/pub/pdb/validation_reports/iq/5iq5 ftp://data.pdbj.org/pub/pdb/validation_reports/iq/5iq5 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 18147.494 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Mayaro virus (strain Brazil) / Details (production host): Gateway cloning system / Production host: Mayaro virus (strain Brazil) / Details (production host): Gateway cloning system / Production host:  References: UniProt: Q8QZ73, Transferases; Transferring one-carbon groups; Methyltransferases, Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases, polynucleotide 5'- ...References: UniProt: Q8QZ73, Transferases; Transferring one-carbon groups; Methyltransferases, Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases, polynucleotide 5'-phosphatase, Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases, nucleoside-triphosphate phosphatase, RNA helicase, RNA-directed RNA polymerase |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
|
|---|
 Movie
Movie Controller
Controller










 PDBj
PDBj

 HSQC
HSQC