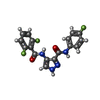
Entry Database : PDB / ID : 5hq0Title Ternary complex of human proteins CDK1, Cyclin B and CKS2, bound to an inhibitor Cyclin-dependent kinase 1 Cyclin-dependent kinases regulatory subunit 2 G2/mitotic-specific cyclin-B1 Keywords / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2.3 Å Authors Noble, M.E. / Martin, M.P. / Korolchuk, S. / Brown, N.R. / Moukhametzianov, R. / Stanley, W.A. Journal : Nat Commun / Year : 2015Title : CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.Authors : Brown, N.R. / Korolchuk, S. / Martin, M.P. / Stanley, W.A. / Moukhametzianov, R. / Noble, M.E. / Endicott, J.A. History Deposition Jan 21, 2016 Deposition site / Processing site Revision 1.0 Mar 2, 2016 Provider / Type Revision 1.1 May 8, 2024 Group / Database references / Category / chem_comp_bond / database_2Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å
MOLECULAR REPLACEMENT / Resolution: 2.3 Å  Authors
Authors Citation
Citation Journal: Nat Commun / Year: 2015
Journal: Nat Commun / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5hq0.cif.gz
5hq0.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5hq0.ent.gz
pdb5hq0.ent.gz PDB format
PDB format 5hq0.json.gz
5hq0.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/hq/5hq0
https://data.pdbj.org/pub/pdb/validation_reports/hq/5hq0 ftp://data.pdbj.org/pub/pdb/validation_reports/hq/5hq0
ftp://data.pdbj.org/pub/pdb/validation_reports/hq/5hq0 Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: CDK1, CDC2, CDC28A, CDKN1, P34CDC2 / Cell line (production host): Sf9 / Production host:
Homo sapiens (human) / Gene: CDK1, CDC2, CDC28A, CDKN1, P34CDC2 / Cell line (production host): Sf9 / Production host: 
 Homo sapiens (human) / Gene: CCNB1, CCNB / Production host:
Homo sapiens (human) / Gene: CCNB1, CCNB / Production host: 
 Homo sapiens (human) / Gene: CKS2 / Production host:
Homo sapiens (human) / Gene: CKS2 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04 / Wavelength: 0.97957 Å
/ Beamline: I04 / Wavelength: 0.97957 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.3→47.26 Å / Cor.coef. Fo:Fc: 0.956 / Cor.coef. Fo:Fc free: 0.917 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.323 / ESU R Free: 0.244
MOLECULAR REPLACEMENT / Resolution: 2.3→47.26 Å / Cor.coef. Fo:Fc: 0.956 / Cor.coef. Fo:Fc free: 0.917 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.323 / ESU R Free: 0.244  Movie
Movie Controller
Controller















 PDBj
PDBj