+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5c9n | ||||||
|---|---|---|---|---|---|---|---|
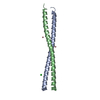
| Title | Crystal structure of GEMC1 coiled-coil domain | ||||||
 Components Components | Geminin coiled-coil domain-containing protein 1 | ||||||
 Keywords Keywords | CELL CYCLE / Geminin Idas coiled coil DNA replication license | ||||||
| Function / homology |  Function and homology information Function and homology informationmulti-ciliated epithelial cell differentiation / cerebrospinal fluid circulation / regulation of DNA-templated DNA replication initiation / seminiferous tubule development / negative regulation of cell cycle / cilium assembly / single fertilization / multicellular organism growth / spermatogenesis / DNA replication ...multi-ciliated epithelial cell differentiation / cerebrospinal fluid circulation / regulation of DNA-templated DNA replication initiation / seminiferous tubule development / negative regulation of cell cycle / cilium assembly / single fertilization / multicellular organism growth / spermatogenesis / DNA replication / chromatin binding / extracellular region / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Caillat, C. / Perrakis, A. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2015 Journal: Acta Crystallogr.,Sect.D / Year: 2015Title: The structure of the GemC1 coiled coil and its interaction with the Geminin family of coiled-coil proteins. Authors: Caillat, C. / Fish, A. / Pefani, D.E. / Taraviras, S. / Lygerou, Z. / Perrakis, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5c9n.cif.gz 5c9n.cif.gz | 71.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5c9n.ent.gz pdb5c9n.ent.gz | 54.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5c9n.json.gz 5c9n.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c9/5c9n https://data.pdbj.org/pub/pdb/validation_reports/c9/5c9n ftp://data.pdbj.org/pub/pdb/validation_reports/c9/5c9n ftp://data.pdbj.org/pub/pdb/validation_reports/c9/5c9n | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1uiiS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 10062.223 Da / Num. of mol.: 2 / Fragment: UNP residues 64-148 / Mutation: L123E, L130E Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GMNC, GEMC1 / Production host: Homo sapiens (human) / Gene: GMNC, GEMC1 / Production host:  #2: Chemical | #3: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.64 Å3/Da / Density % sol: 66.24 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: 0.1 M Hepes buffer pH 7.5, 7% Ethanol, 10% 2-Methyl-2,4-pentanediol (MPD) 0.01 M Ethylene-diamine-tetra-acetic acid disodium salt dihydrate. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-2 / Wavelength: 0.8726 Å / Beamline: ID23-2 / Wavelength: 0.8726 Å |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Jul 3, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.8726 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→53.8 Å / Num. obs: 15376 / % possible obs: 99.5 % / Redundancy: 3.7 % / Rmerge(I) obs: 0.07 / Net I/σ(I): 11.8 |
| Reflection shell | Resolution: 2.2→2.32 Å / Redundancy: 3.8 % / Rmerge(I) obs: 0.871 / Mean I/σ(I) obs: 1.5 / % possible all: 99.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1UII Resolution: 2.2→53.78 Å / Cor.coef. Fo:Fc: 0.95 / Cor.coef. Fo:Fc free: 0.945 / SU B: 11.959 / SU ML: 0.14 / Cross valid method: THROUGHOUT / ESU R: 0.175 / ESU R Free: 0.16 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.7 Å / Shrinkage radii: 0.7 Å / VDW probe radii: 1 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 54.241 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→53.78 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj

