[English] 日本語
 Yorodumi
Yorodumi- PDB-4xp5: X-ray structure of Drosophila dopamine transporter bound to cocai... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4xp5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | X-ray structure of Drosophila dopamine transporter bound to cocaine analogue-RTI55 | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | transport protein/inhibitor / integral membrane protein / membrane protein-transport protein complex / transport protein-inhibitor complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationcatecholamine uptake / : / SLC-mediated transport of neurotransmitters / circadian sleep/wake cycle / sodium:chloride symporter activity / response to odorant / cocaine binding / norepinephrine transport / dopamine:sodium symporter activity / regulation of presynaptic cytosolic calcium ion concentration ...catecholamine uptake / : / SLC-mediated transport of neurotransmitters / circadian sleep/wake cycle / sodium:chloride symporter activity / response to odorant / cocaine binding / norepinephrine transport / dopamine:sodium symporter activity / regulation of presynaptic cytosolic calcium ion concentration / dopamine transport / monoamine transmembrane transporter activity / sleep / neuronal cell body membrane / neurotransmitter transport / dopamine uptake involved in synaptic transmission / amino acid transport / sodium ion transmembrane transport / adult locomotory behavior / presynaptic membrane / axon / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 3.3 Å SYNCHROTRON / Resolution: 3.3 Å | |||||||||
 Authors Authors | Gouaux, E. / Penmatsa, A. / Wang, K. | |||||||||
 Citation Citation |  Journal: Nature / Year: 2015 Journal: Nature / Year: 2015Title: Neurotransmitter and psychostimulant recognition by the dopamine transporter. Authors: Wang, K.H. / Penmatsa, A. / Gouaux, E. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4xp5.cif.gz 4xp5.cif.gz | 205.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4xp5.ent.gz pdb4xp5.ent.gz | 158.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4xp5.json.gz 4xp5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xp/4xp5 https://data.pdbj.org/pub/pdb/validation_reports/xp/4xp5 ftp://data.pdbj.org/pub/pdb/validation_reports/xp/4xp5 ftp://data.pdbj.org/pub/pdb/validation_reports/xp/4xp5 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4xp1C  4xp4C  4xp6C  4xp9C  4xpaC  4xpbC  4xpfC  4xpgC  4xphC  4xptC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | biological unit is the same as asym. |
- Components
Components
-Antibody , 2 types, 2 molecules LH
| #2: Antibody | Mass: 23177.473 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #3: Antibody | Mass: 23619.430 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein / Sugars , 2 types, 2 molecules A
| #1: Protein | Mass: 59892.289 Da / Num. of mol.: 1 / Mutation: V74A, L415A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: A0A0B4KEX2, UniProt: Q7K4Y6*PLUS Homo sapiens (human) / References: UniProt: A0A0B4KEX2, UniProt: Q7K4Y6*PLUS |
|---|---|
| #4: Polysaccharide | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose / alpha-maltose |
-Non-polymers , 5 types, 7 molecules 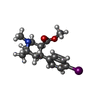








| #5: Chemical | ChemComp-42F / | ||||||
|---|---|---|---|---|---|---|---|
| #6: Chemical | | #7: Chemical | ChemComp-P4G / | #8: Chemical | #9: Chemical | ChemComp-CL / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.03 Å3/Da / Density % sol: 75.56 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / Details: PEG 400 (41%), Tris (0.1M) / PH range: pH 8.0 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1 Å / Beamline: 5.0.2 / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Feb 22, 2014 |
| Radiation | Monochromator: Si111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3.3→50 Å / Num. obs: 28495 / % possible obs: 86.2 % / Redundancy: 4.3 % / Net I/σ(I): 11 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 3.3→47.28 Å / SU ML: 0.44 / Cross valid method: FREE R-VALUE / σ(F): 1.36 / Phase error: 29.96 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 151.69 Å2 / Biso mean: 72.9476 Å2 / Biso min: 20 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.3→47.28 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




















