| 登録情報 | データベース: PDB / ID: 4xak
|
|---|
| タイトル | Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD |
|---|
 要素 要素 | - Heavy chain of neutralizing antibody m336
- Light chain of neutralizing antibody m336
- Spike glycoprotein
|
|---|
 キーワード キーワード | IMMUNE SYSTEM / MERS-CoV RBD / antibody m336 / neutralization |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane類似検索 - 分子機能 ubp-family deubiquitinating enzyme fold - #30 / Spike protein, C-terminal core receptor binding subdomain / ubp-family deubiquitinating enzyme fold / Spike (S) protein S1 subunit, receptor-binding domain, MERS-CoV / Spike (S) protein S1 subunit, N-terminal domain, MERS-CoV-like / Spike glycoprotein S2, coronavirus, C-terminal / Coronavirus spike glycoprotein S2, intravirion / Single Sheet / Coronavirus spike glycoprotein S1, C-terminal / Coronavirus spike glycoprotein S1, C-terminal ...ubp-family deubiquitinating enzyme fold - #30 / Spike protein, C-terminal core receptor binding subdomain / ubp-family deubiquitinating enzyme fold / Spike (S) protein S1 subunit, receptor-binding domain, MERS-CoV / Spike (S) protein S1 subunit, N-terminal domain, MERS-CoV-like / Spike glycoprotein S2, coronavirus, C-terminal / Coronavirus spike glycoprotein S2, intravirion / Single Sheet / Coronavirus spike glycoprotein S1, C-terminal / Coronavirus spike glycoprotein S1, C-terminal / Spike glycoprotein, N-terminal domain superfamily / Spike S1 subunit, receptor binding domain superfamily, betacoronavirus / Spike glycoprotein, betacoronavirus / Betacoronavirus spike (S) glycoprotein S1 subunit N-terminal (NTD) domain profile. / Betacoronavirus spike (S) glycoprotein S1 subunit C-terminal (CTD) domain profile. / Spike (S) protein S1 subunit, receptor-binding domain, betacoronavirus / Betacoronavirus spike glycoprotein S1, receptor binding / Spike glycoprotein S1, N-terminal domain, betacoronavirus-like / Betacoronavirus-like spike glycoprotein S1, N-terminal / Spike glycoprotein S2 superfamily, coronavirus / Spike glycoprotein S2, coronavirus, heptad repeat 1 / Spike glycoprotein S2, coronavirus, heptad repeat 2 / Coronavirus spike (S) glycoprotein S2 subunit heptad repeat 1 (HR1) region profile. / Coronavirus spike (S) glycoprotein S2 subunit heptad repeat 2 (HR2) region profile. / Spike glycoprotein S2, coronavirus / Coronavirus spike glycoprotein S2 / Alpha-Beta Plaits / Immunoglobulins / Immunoglobulin-like / Sandwich / 2-Layer Sandwich / Mainly Beta / Alpha Beta類似検索 - ドメイン・相同性 |
|---|
| 生物種 |  Human coronavirus EMC (ウイルス) Human coronavirus EMC (ウイルス)
 Homo sapiens (ヒト) Homo sapiens (ヒト) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.45 Å 分子置換 / 解像度: 2.45 Å |
|---|
 データ登録者 データ登録者 | Zhou, T. / Dimtrov, D.S. / Ying, T. |
|---|
 引用 引用 |  ジャーナル: Nat Commun / 年: 2015 ジャーナル: Nat Commun / 年: 2015
タイトル: Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
著者: Ying, T. / Prabakaran, P. / Du, L. / Shi, W. / Feng, Y. / Wang, Y. / Wang, L. / Li, W. / Jiang, S. / Dimitrov, D.S. / Zhou, T. |
|---|
| 履歴 | | 登録 | 2014年12月15日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2015年8月26日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2015年9月23日 | Group: Database references |
|---|
| 改定 1.2 | 2017年11月22日 | Group: Derived calculations / Refinement description / カテゴリ: pdbx_struct_oper_list / software / Item: _pdbx_struct_oper_list.symmetry_operation |
|---|
| 改定 1.3 | 2020年7月29日 | Group: Data collection / Derived calculations / Structure summary
カテゴリ: chem_comp / entity ...chem_comp / entity / pdbx_chem_comp_identifier / pdbx_entity_nonpoly / struct_conn / struct_site / struct_site_gen
Item: _chem_comp.name / _chem_comp.type ..._chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_entity_nonpoly.name / _struct_conn.pdbx_role
解説: Carbohydrate remediation / Provider: repository / タイプ: Remediation |
|---|
| 改定 1.4 | 2023年9月27日 | Group: Data collection / Database references ...Data collection / Database references / Refinement description / Structure summary
カテゴリ: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| 改定 1.5 | 2024年10月30日 | Group: Structure summary
カテゴリ: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Human coronavirus EMC (ウイルス)
Human coronavirus EMC (ウイルス) Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.45 Å
分子置換 / 解像度: 2.45 Å  データ登録者
データ登録者 引用
引用 ジャーナル: Nat Commun / 年: 2015
ジャーナル: Nat Commun / 年: 2015 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 4xak.cif.gz
4xak.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb4xak.ent.gz
pdb4xak.ent.gz PDB形式
PDB形式 4xak.json.gz
4xak.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 4xak_validation.pdf.gz
4xak_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 4xak_full_validation.pdf.gz
4xak_full_validation.pdf.gz 4xak_validation.xml.gz
4xak_validation.xml.gz 4xak_validation.cif.gz
4xak_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/xa/4xak
https://data.pdbj.org/pub/pdb/validation_reports/xa/4xak ftp://data.pdbj.org/pub/pdb/validation_reports/xa/4xak
ftp://data.pdbj.org/pub/pdb/validation_reports/xa/4xak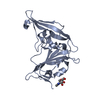
 リンク
リンク 集合体
集合体


 要素
要素 Homo sapiens (ヒト) / 細胞株 (発現宿主): HEK 293F / 発現宿主:
Homo sapiens (ヒト) / 細胞株 (発現宿主): HEK 293F / 発現宿主:  Homo sapiens (ヒト)
Homo sapiens (ヒト) Homo sapiens (ヒト) / プラスミド: plasmid / 細胞株 (発現宿主): HEK 293F / 発現宿主:
Homo sapiens (ヒト) / プラスミド: plasmid / 細胞株 (発現宿主): HEK 293F / 発現宿主:  Homo sapiens (ヒト)
Homo sapiens (ヒト)

 Human coronavirus EMC (ウイルス) / 株: isolate United Kingdom/H123990006/2012 / 遺伝子: S, 3 / プラスミド: pVRC8400 / 細胞株 (発現宿主): GNTI-/- / 発現宿主:
Human coronavirus EMC (ウイルス) / 株: isolate United Kingdom/H123990006/2012 / 遺伝子: S, 3 / プラスミド: pVRC8400 / 細胞株 (発現宿主): GNTI-/- / 発現宿主:  Homo sapiens (ヒト) / 参照: UniProt: K9N5Q8
Homo sapiens (ヒト) / 参照: UniProt: K9N5Q8


 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 22-ID / 波長: 1 Å
/ ビームライン: 22-ID / 波長: 1 Å 分子置換
分子置換 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー













 PDBj
PDBj


