Entry Database : PDB / ID : 4xakTitle Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD Heavy chain of neutralizing antibody m336 Light chain of neutralizing antibody m336 Spike glycoprotein Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / / Resolution : 2.45 Å Authors Zhou, T. / Dimtrov, D.S. / Ying, T. Journal : Nat Commun / Year : 2015Title : Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.Authors : Ying, T. / Prabakaran, P. / Du, L. / Shi, W. / Feng, Y. / Wang, Y. / Wang, L. / Li, W. / Jiang, S. / Dimitrov, D.S. / Zhou, T. History Deposition Dec 15, 2014 Deposition site / Processing site Revision 1.0 Aug 26, 2015 Provider / Type Revision 1.1 Sep 23, 2015 Group Revision 1.2 Nov 22, 2017 Group / Refinement description / Category / software / Item Revision 1.3 Jul 29, 2020 Group / Derived calculations / Structure summaryCategory chem_comp / entity ... chem_comp / entity / pdbx_chem_comp_identifier / pdbx_entity_nonpoly / struct_conn / struct_site / struct_site_gen Item _chem_comp.name / _chem_comp.type ... _chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_entity_nonpoly.name / _struct_conn.pdbx_role Description / Provider / Type Revision 1.4 Sep 27, 2023 Group Data collection / Database references ... Data collection / Database references / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_DOI / _database_2.pdbx_database_accessionRevision 1.5 Oct 30, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Human coronavirus EMC
Human coronavirus EMC Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.45 Å
molecular replacement / Resolution: 2.45 Å  Authors
Authors Citation
Citation Journal: Nat Commun / Year: 2015
Journal: Nat Commun / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4xak.cif.gz
4xak.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4xak.ent.gz
pdb4xak.ent.gz PDB format
PDB format 4xak.json.gz
4xak.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/xa/4xak
https://data.pdbj.org/pub/pdb/validation_reports/xa/4xak ftp://data.pdbj.org/pub/pdb/validation_reports/xa/4xak
ftp://data.pdbj.org/pub/pdb/validation_reports/xa/4xak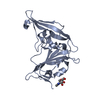
 Links
Links Assembly
Assembly


 Components
Components Homo sapiens (human) / Cell line (production host): HEK 293F / Production host:
Homo sapiens (human) / Cell line (production host): HEK 293F / Production host:  Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human) / Plasmid: plasmid / Cell line (production host): HEK 293F / Production host:
Homo sapiens (human) / Plasmid: plasmid / Cell line (production host): HEK 293F / Production host:  Homo sapiens (human)
Homo sapiens (human)

 Human coronavirus EMC / Strain: isolate United Kingdom/H123990006/2012 / Gene: S, 3 / Plasmid: pVRC8400 / Cell line (production host): GNTI-/- / Production host:
Human coronavirus EMC / Strain: isolate United Kingdom/H123990006/2012 / Gene: S, 3 / Plasmid: pVRC8400 / Cell line (production host): GNTI-/- / Production host:  Homo sapiens (human) / References: UniProt: K9N5Q8
Homo sapiens (human) / References: UniProt: K9N5Q8


 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 22-ID / Wavelength: 1 Å
/ Beamline: 22-ID / Wavelength: 1 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj


