[English] 日本語
 Yorodumi
Yorodumi- PDB-4x20: Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4x20 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | ||||||
 Components Components |
| ||||||
 Keywords Keywords | Structural Protein/Inhibitor / Binding Sites / Competitive / Cattle / Tumor / Colchicine / Humans / Microtubules / Protein Binding / Protein Conformation / Protein Multimerization / Tubulin / Tubulin Modulators / Structural Protein-Inhibitor complex | ||||||
| Function / homology |  Function and homology information Function and homology informationaxonemal microtubule / organelle transport along microtubule / forebrain morphogenesis / cerebellar cortex morphogenesis / glial cell differentiation / dentate gyrus development / neuron projection arborization / flagellated sperm motility / pyramidal neuron differentiation / response to L-glutamate ...axonemal microtubule / organelle transport along microtubule / forebrain morphogenesis / cerebellar cortex morphogenesis / glial cell differentiation / dentate gyrus development / neuron projection arborization / flagellated sperm motility / pyramidal neuron differentiation / response to L-glutamate / microtubule depolymerization / centrosome cycle / smoothened signaling pathway / regulation of synapse organization / startle response / motor behavior / response to tumor necrosis factor / microtubule polymerization / locomotory exploration behavior / regulation of microtubule polymerization or depolymerization / response to mechanical stimulus / sperm flagellum / microtubule-based process / condensed chromosome / homeostasis of number of cells within a tissue / cellular response to calcium ion / tubulin binding / adult locomotory behavior / intracellular protein transport / neuromuscular junction / recycling endosome / synapse organization / cerebral cortex development / visual learning / structural constituent of cytoskeleton / memory / neuron migration / neuron projection development / cytoplasmic ribonucleoprotein granule / growth cone / neuron apoptotic process / gene expression / microtubule / neuron projection / protein heterodimerization activity / hydrolase activity / GTPase activity / GTP binding / protein-containing complex binding / Golgi apparatus / metal ion binding / identical protein binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |   | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.5 Å MOLECULAR REPLACEMENT / Resolution: 3.5 Å | ||||||
 Authors Authors | Parris, K.D. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2014 Journal: J.Med.Chem. / Year: 2014Title: Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications. Authors: Maderna, A. / Doroski, M. / Subramanyam, C. / Porte, A. / Leverett, C.A. / Vetelino, B.C. / Chen, Z. / Risley, H. / Parris, K. / Pandit, J. / Varghese, A.H. / Shanker, S. / Song, C. / ...Authors: Maderna, A. / Doroski, M. / Subramanyam, C. / Porte, A. / Leverett, C.A. / Vetelino, B.C. / Chen, Z. / Risley, H. / Parris, K. / Pandit, J. / Varghese, A.H. / Shanker, S. / Song, C. / Sukuru, S.C. / Farley, K.A. / Wagenaar, M.M. / Shapiro, M.J. / Musto, S. / Lam, M.H. / Loganzo, F. / O'Donnell, C.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4x20.cif.gz 4x20.cif.gz | 383.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4x20.ent.gz pdb4x20.ent.gz | 303.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4x20.json.gz 4x20.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/x2/4x20 https://data.pdbj.org/pub/pdb/validation_reports/x2/4x20 ftp://data.pdbj.org/pub/pdb/validation_reports/x2/4x20 ftp://data.pdbj.org/pub/pdb/validation_reports/x2/4x20 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4x1iC  4x1kC  4x1yC  3hkbS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 3 types, 5 molecules ACBDE
| #1: Protein | Mass: 50188.441 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #2: Protein | Mass: 49969.797 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #3: Protein | | Mass: 16719.938 Da / Num. of mol.: 1 / Fragment: UNP residues 49-189 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 10 molecules 


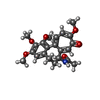
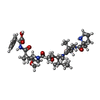




| #4: Chemical | | #5: Chemical | #6: Chemical | #7: Chemical | #8: Chemical |   Type: peptide-like, Peptide-like / Class: Inhibitor / Mass: 729.946 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C39H63N5O8 Type: peptide-like, Peptide-like / Class: Inhibitor / Mass: 729.946 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C39H63N5O8References: 2-methyl-L-prolyl-N-[(3R,4S,5S)-1-{(2S)-2-[(1R,2R)-3-{[(1S)-1-carboxy-2-phenylethyl]amino}-1-methoxy-2-methyl-3-oxopropyl]pyrrolidin-1-yl}-3-methoxy-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.62 Å3/Da / Density % sol: 53.05 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6.8 Details: 5% PEG 400, 0.1M LiSO4, 5-7% PEG 20K, 50 mM K-PIPES |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 Å / Beamline: 17-ID / Wavelength: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Feb 9, 2012 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 3.5→50 Å / Num. obs: 25405 / % possible obs: 90.1 % / Redundancy: 4 % / Biso Wilson estimate: 106.47 Å2 / Rmerge(I) obs: 0.12 / Χ2: 0.821 / Net I/av σ(I): 11.087 / Net I/σ(I): 3.7 / Num. measured all: 101790 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3HKB Resolution: 3.5→40.83 Å / Cor.coef. Fo:Fc: 0.9136 / Cor.coef. Fo:Fc free: 0.8925 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.682
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 147.36 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.798 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.5→40.83 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.5→3.64 Å / Total num. of bins used: 13
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj






