[English] 日本語
 Yorodumi
Yorodumi- PDB-4uyo: Structure of delta7-DgkA in 7.9 MAG by serial femtosecond crystat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4uyo | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of delta7-DgkA in 7.9 MAG by serial femtosecond crystatallography to 2.18 angstrom resolution | ||||||
 Components Components | DIACYLGLYCEROL KINASE-DELTA 7 | ||||||
 Keywords Keywords | TRANSFERASE / 7.9 MAG / IN MESO CRYSTALLIZATION / LIPID CUBIC PHASE / LIPIDIC CUBIC PHASE / LIPID MESOPHASE / LIPIDIC MESOPHASE / MEMBRANE PROTEIN / MICROCRYSTALS / MONOACYLGLYCEROL / ROOM TEMPERATURE CRYSTALLOGRAPHY / SERIAL FEMTOSECOND CRYSTALLOGRAPHY / X-RAY FREE-ELECTRON LASER | ||||||
| Function / homology |  Function and homology information Function and homology informationdiacylglycerol kinase (ATP) / lipid kinase activity / ATP-dependent diacylglycerol kinase activity / phosphatidic acid biosynthetic process / response to UV / ATP binding / metal ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  FREE ELECTRON LASER / FREE ELECTRON LASER /  MOLECULAR REPLACEMENT / Resolution: 2.18 Å MOLECULAR REPLACEMENT / Resolution: 2.18 Å | ||||||
 Authors Authors | Li, D. / Howe, N. / Other, O. / Caffrey, M. | ||||||
 Citation Citation |  Journal: Nat.Commun. / Year: 2015 Journal: Nat.Commun. / Year: 2015Title: Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase. Authors: Li, D. / Stansfeld, P.J. / Sansom, M.S.P. / Keogh, A. / Vogeley, L. / Howe, N. / Lyons, J.A. / Aragao, D. / Fromme, P. / Fromme, R. / Basu, S. / Grotjohann, I. / Kupitz, C. / Rendek, K. / ...Authors: Li, D. / Stansfeld, P.J. / Sansom, M.S.P. / Keogh, A. / Vogeley, L. / Howe, N. / Lyons, J.A. / Aragao, D. / Fromme, P. / Fromme, R. / Basu, S. / Grotjohann, I. / Kupitz, C. / Rendek, K. / Weierstall, U. / Zatsepin, N.A. / Cherezov, V. / Liu, W. / Bandaru, S. / English, N.J. / Gati, C. / Barty, A. / Yefanov, O. / Chapman, H.N. / Diederichs, K. / Messerschmidt, M. / Boutet, S. / Williams, G.J. / Marvin Seibert, M. / Caffrey, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4uyo.cif.gz 4uyo.cif.gz | 142.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4uyo.ent.gz pdb4uyo.ent.gz | 113.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4uyo.json.gz 4uyo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uy/4uyo https://data.pdbj.org/pub/pdb/validation_reports/uy/4uyo ftp://data.pdbj.org/pub/pdb/validation_reports/uy/4uyo ftp://data.pdbj.org/pub/pdb/validation_reports/uy/4uyo | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4uxwC  4uxxC  4uxzC  3ze3S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 6 molecules ABCDEF
| #1: Protein | Mass: 14204.451 Da / Num. of mol.: 6 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 96 molecules 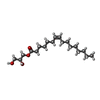








| #2: Chemical | ChemComp-79N / ( #3: Chemical | ChemComp-79M / ( #4: Chemical | ChemComp-ZN / | #5: Chemical | ChemComp-FLC / | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 140440 X-RAY DIFFRACTION / Number of used crystals: 140440 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.93 Å3/Da / Density % sol: 58.04 % / Description: R SPLIT (I) 0.07, R SPLIT FOR SHELL (I) 2.38 |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase / pH: 5.6 Details: 0.2 %(V/V) 2-METHYL-2, 4-PENTANEDIOL (MPD), 0.1 M SODIUM CHLORIDE, 0.1 M SODIUM CITRATE/HCL PH 5.6. CRYSTALLIZED USING THE IN MESO (LIPID CUBIC PHASE) METHOD AT 20 DEGREES CELCIUS WITH THE 7. ...Details: 0.2 %(V/V) 2-METHYL-2, 4-PENTANEDIOL (MPD), 0.1 M SODIUM CHLORIDE, 0.1 M SODIUM CITRATE/HCL PH 5.6. CRYSTALLIZED USING THE IN MESO (LIPID CUBIC PHASE) METHOD AT 20 DEGREES CELCIUS WITH THE 7.9 MONOACYLGLYCEROL (7.9 MAG) AS THE HOSTING LIPID.CRYSTALLIZATION WAS SET UP BY INCUBATING 20 MICROLITERS OF LIPID CUBIC PHASE WITH 400 MICROLITERS OF PRECIPITANT SOLUTION IN COUPLED SYRINGES. |
-Data collection
| Diffraction | Mean temperature: 293 K / Serial crystal experiment: Y |
|---|---|
| Diffraction source | Source:  FREE ELECTRON LASER / Site: FREE ELECTRON LASER / Site:  SLAC LCLS SLAC LCLS  / Beamline: CXI / Wavelength: 1.302 / Wavelength: 1.302 Å / Beamline: CXI / Wavelength: 1.302 / Wavelength: 1.302 Å |
| Detector | Type: Cornell-SLAC Pixel Array Detector (CSPAD) / Detector: PIXEL / Date: Mar 25, 2013 |
| Radiation | Monochromator: K-B MIRRORS / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.302 Å / Relative weight: 1 |
| Reflection | Resolution: 2.18→40.5 Å / Num. obs: 54058 / % possible obs: 100 % / Observed criterion σ(I): -5 / Redundancy: 3466 % / Biso Wilson estimate: 71.31 Å2 / Net I/σ(I): 19 |
| Reflection shell | Resolution: 2.18→2.24 Å / Redundancy: 3519 % / Mean I/σ(I) obs: 0.44 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3ZE3 Resolution: 2.18→40.013 Å / SU ML: 0.32 / σ(F): 1.33 / Phase error: 28.73 / Stereochemistry target values: ML Details: THERE ARE SIX NCS-RELATED MOLECULES IN THE ASYMMETRIC UNIT BUT NCS RESTRAINTS WERE NOT USED IN THE REFINEMENT.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 70.42 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.18→40.013 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj









