+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4uuz | ||||||
|---|---|---|---|---|---|---|---|
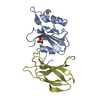
| Title | MCM2-histone complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN / DNA-BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationHDMs demethylate histones / PKMTs methylate histone lysines / Interleukin-7 signaling / Chromatin modifying enzymes / : / SUMOylation of chromatin organization proteins / Factors involved in megakaryocyte development and platelet production / RCAF complex / RMTs methylate histone arginines / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks ...HDMs demethylate histones / PKMTs methylate histone lysines / Interleukin-7 signaling / Chromatin modifying enzymes / : / SUMOylation of chromatin organization proteins / Factors involved in megakaryocyte development and platelet production / RCAF complex / RMTs methylate histone arginines / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / SIRT1 negatively regulates rRNA expression / NoRC negatively regulates rRNA expression / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / Formation of the beta-catenin:TCF transactivating complex / PRC2 methylates histones and DNA / HDACs deacetylate histones / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / RNA Polymerase I Promoter Escape / Regulation of endogenous retroelements by KRAB-ZFP proteins / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / Senescence-Associated Secretory Phenotype (SASP) / Transcriptional regulation by small RNAs / Estrogen-dependent gene expression / HATs acetylate histones / Assembly of the ORC complex at the origin of replication / Oxidative Stress Induced Senescence / Switching of origins to a post-replicative state / Unwinding of DNA / polytene chromosome / nuclear origin of replication recognition complex / Regulation of MITF-M-dependent genes involved in DNA replication, damage repair and senescence / CMG complex / MCM complex / double-strand break repair via break-induced replication / mitotic DNA replication initiation / regulation of DNA-templated DNA replication initiation / nucleosomal DNA binding / nuclear chromosome / DNA replication origin binding / cochlea development / DNA replication initiation / Activation of the pre-replicative complex / Activation of ATR in response to replication stress / cellular response to interleukin-4 / DNA helicase activity / Assembly of the pre-replicative complex / Orc1 removal from chromatin / structural constituent of chromatin / nucleosome / nucleosome assembly / single-stranded DNA binding / chromosome / histone binding / DNA helicase / DNA replication / chromosome, telomeric region / cilium / protein heterodimerization activity / intracellular membrane-bounded organelle / apoptotic process / chromatin / enzyme binding / ATP hydrolysis activity / DNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |   HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å | ||||||
 Authors Authors | Richet, N. / Liu, D. / Legrand, P. / Bakail, M. / Compper, C. / Besle, A. / Guerois, R. / Ochsenbein, F. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2015 Journal: Nucleic Acids Res. / Year: 2015Title: Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork. Authors: Richet, N. / Liu, D. / Legrand, P. / Velours, C. / Corpet, A. / Gaubert, A. / Bakail, M. / Moal-Raisin, G. / Guerois, R. / Compper, C. / Besle, A. / Guichard, B. / Almouzni, G. / Ochsenbein, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4uuz.cif.gz 4uuz.cif.gz | 100.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4uuz.ent.gz pdb4uuz.ent.gz | 78.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4uuz.json.gz 4uuz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4uuz_validation.pdf.gz 4uuz_validation.pdf.gz | 441.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4uuz_full_validation.pdf.gz 4uuz_full_validation.pdf.gz | 443.2 KB | Display | |
| Data in XML |  4uuz_validation.xml.gz 4uuz_validation.xml.gz | 9.6 KB | Display | |
| Data in CIF |  4uuz_validation.cif.gz 4uuz_validation.cif.gz | 12 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uu/4uuz https://data.pdbj.org/pub/pdb/validation_reports/uu/4uuz ftp://data.pdbj.org/pub/pdb/validation_reports/uu/4uuz ftp://data.pdbj.org/pub/pdb/validation_reports/uu/4uuz | HTTPS FTP |
-Related structure data
| Related structure data |  1kx5S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 15421.101 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein | Mass: 11408.452 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #3: Protein | Mass: 7968.699 Da / Num. of mol.: 1 / Fragment: RESIDUES 69-138 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PETM30 / Production host: HOMO SAPIENS (human) / Plasmid: PETM30 / Production host:  |
| Sequence details | FRAGMENT FROM AMINO-ACID 69 TO 138 |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density % sol: 46.7 % Description: SULFUR ATOM POSITIONS WERE CONFIRMED BY ANALYZING THE ANOMALOUS DIFFERENCE FOURIER MAPS CALCULATED WITH THE PROGRAM ANODE. |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7 / Details: 0.1M HEPES PH7, 21% PEG3000 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 1.65312 / Beamline: PROXIMA 1 / Wavelength: 1.65312 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jul 5, 2014 / Details: KIRKPATRICK-BAEZ PAIR OF BI-MORPH MIRRORS |
| Radiation | Monochromator: SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.65312 Å / Relative weight: 1 |
| Reflection | Resolution: 2.9→60 Å / Num. obs: 5602 / % possible obs: 99.9 % / Observed criterion σ(I): -3 / Redundancy: 10.8 % / Biso Wilson estimate: 159.78 Å2 / Rmerge(I) obs: 0.08 / Net I/σ(I): 19.7 |
| Reflection shell | Resolution: 2.9→2.97 Å / Redundancy: 10.5 % / Rmerge(I) obs: 1.5 / Mean I/σ(I) obs: 1.2 / % possible all: 98.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1KX5 Resolution: 2.9→57.89 Å / Cor.coef. Fo:Fc: 0.9502 / Cor.coef. Fo:Fc free: 0.9298 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.355 Details: IDEAL-DIST CONTACT TERM CONTACT SETUP. ALL ATOMS HAVE CCP4 ATOM TYPE FROM LIBRARY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 131.95 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.716 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→57.89 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.9→3.24 Å / Total num. of bins used: 5
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj





