[English] 日本語
 Yorodumi
Yorodumi- PDB-4u3d: LpxC from A.Aaeolicus in complex with 4-[[4-[2-[4-(morpholinometh... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4u3d | ||||||
|---|---|---|---|---|---|---|---|
| Title | LpxC from A.Aaeolicus in complex with 4-[[4-[2-[4-(morpholinomethyl)phenyl]ethynyl]phenoxy]methyl]tetrahydropyran-4-carbohydroxamic acid (compound 9) | ||||||
 Components Components | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase | ||||||
 Keywords Keywords | HYDROLASE / antibacterial / lpxc / gram negative bacteria / MMP / hydrophobe | ||||||
| Function / homology |  Function and homology information Function and homology informationUDP-3-O-acyl-N-acetylglucosamine deacetylase / UDP-3-O-acyl-N-acetylglucosamine deacetylase activity / lipid A biosynthetic process / metal ion binding / membrane Similarity search - Function | ||||||
| Biological species |   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.25 Å MOLECULAR REPLACEMENT / Resolution: 1.25 Å | ||||||
 Authors Authors | Olivier, N.B. | ||||||
 Citation Citation |  Journal: Acs Med.Chem.Lett. / Year: 2014 Journal: Acs Med.Chem.Lett. / Year: 2014Title: Synthesis, Structure, and SAR of Tetrahydropyran-Based LpxC Inhibitors. Authors: Murphy-Benenato, K.E. / Olivier, N. / Choy, A. / Ross, P.L. / Miller, M.D. / Thresher, J. / Gao, N. / Hale, M.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4u3d.cif.gz 4u3d.cif.gz | 83.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4u3d.ent.gz pdb4u3d.ent.gz | 59.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4u3d.json.gz 4u3d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/u3/4u3d https://data.pdbj.org/pub/pdb/validation_reports/u3/4u3d ftp://data.pdbj.org/pub/pdb/validation_reports/u3/4u3d ftp://data.pdbj.org/pub/pdb/validation_reports/u3/4u3d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4u3bC  2go4S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | biological unit is the same as asym. |
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 30989.510 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Aquifex aeolicus (bacteria) / Strain: VF5 / Gene: lpxC, envA, aq_1772 / Production host: Aquifex aeolicus (bacteria) / Strain: VF5 / Gene: lpxC, envA, aq_1772 / Production host:  References: UniProt: O67648, Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides |
|---|
-Non-polymers , 5 types, 382 molecules 


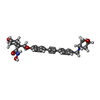





| #2: Chemical | ChemComp-IMD / | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | | #4: Chemical | ChemComp-CL / | #5: Chemical | ChemComp-3BX / | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.69 Å3/Da / Density % sol: 54.33 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 6.4 Details: Protein solution: 25 mM Tris-HCl, pH 8.0, 0.15 M NaCl, 0.1 mM ZnSO4, 5% glycerol, 1 mM inhibitor. Well solution: 15% PEG 550 MME, 15% PEG 20K, 50 mM Imidazole and 50 mM MES (Morpheus buffer ...Details: Protein solution: 25 mM Tris-HCl, pH 8.0, 0.15 M NaCl, 0.1 mM ZnSO4, 5% glycerol, 1 mM inhibitor. Well solution: 15% PEG 550 MME, 15% PEG 20K, 50 mM Imidazole and 50 mM MES (Morpheus buffer 1), pH 6.5, 2% 1,6-Hexanediol; 2% 1-Butanol 1,2-Propanediol (racemic); 2% 2-Propanol; 2% 1,4-Butanediol; 2% 1,3-Propanediol PH range: 6.5-7.5 |
-Data collection
| Diffraction | Mean temperature: 110 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 Å / Beamline: 17-ID / Wavelength: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jul 16, 2012 / Details: monochromator | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Si 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.246→133.282 Å / Num. all: 89070 / Num. obs: 89070 / % possible obs: 97.8 % / Redundancy: 10.8 % / Biso Wilson estimate: 12.26 Å2 / Rpim(I) all: 0.024 / Rrim(I) all: 0.08 / Rsym value: 0.071 / Net I/av σ(I): 5.763 / Net I/σ(I): 20.9 / Num. measured all: 958795 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2GO4 Resolution: 1.25→35.05 Å / Cor.coef. Fo:Fc: 0.9677 / Cor.coef. Fo:Fc free: 0.9646 / SU R Cruickshank DPI: 0.037 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.038 / SU Rfree Blow DPI: 0.038 / SU Rfree Cruickshank DPI: 0.037
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 122.8 Å2 / Biso mean: 18.21 Å2 / Biso min: 6.55 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.121 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.25→35.05 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.25→1.28 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








