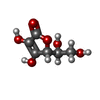
Entry Database : PDB / ID : 4twlTitle Crystal structure of dioscorin complexed with ascorbate Dioscorin 5 Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Dioscorea japonica (Japanese yam)Method / / Resolution : 2.11 Å Authors Xue, Y.L. / Miyakawa, T. / Nakamura, A. / Tanokura, M. Funding support Organization Grant number Country High Energy Accelerator Research Organization 2011G605 Targeted Proteins Research Program (TPRP) of the Ministry of Education, Culture, Sports, Science and Technology of Japan National Natural Science Foundation of China 31201285 Scientific Research Foundation for the Returned Overseas Chinese Scholars 2013693
Journal : Mol Plant / Year : 2015Title : Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate ReductaseAuthors : Xue, Y.L. / Miyakawa, T. / Nakamura, A. / Hatano, K. / Sawano, Y. / Tanokura, M. History Deposition Jul 1, 2014 Deposition site / Processing site Revision 1.0 Apr 1, 2015 Provider / Type Revision 1.1 Aug 5, 2015 Group Revision 1.2 Jan 29, 2020 Group / Derived calculations / Category / pdbx_struct_oper_listItem / _pdbx_struct_oper_list.symmetry_operationRevision 1.3 Jul 29, 2020 Group / Derived calculations / Category / struct_site / struct_site_gen / Item / Description / Provider / Type Revision 1.4 Oct 30, 2024 Group / Database references / Structure summaryCategory chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature Item / _database_2.pdbx_DOI / _database_2.pdbx_database_accession
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Dioscorea japonica (Japanese yam)
Dioscorea japonica (Japanese yam) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.11 Å
SYNCHROTRON / Resolution: 2.11 Å  Authors
Authors Japan,
Japan,  China, 4items
China, 4items  Citation
Citation Journal: Mol Plant / Year: 2015
Journal: Mol Plant / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4twl.cif.gz
4twl.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4twl.ent.gz
pdb4twl.ent.gz PDB format
PDB format 4twl.json.gz
4twl.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 4twl_validation.pdf.gz
4twl_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 4twl_full_validation.pdf.gz
4twl_full_validation.pdf.gz 4twl_validation.xml.gz
4twl_validation.xml.gz 4twl_validation.cif.gz
4twl_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/tw/4twl
https://data.pdbj.org/pub/pdb/validation_reports/tw/4twl ftp://data.pdbj.org/pub/pdb/validation_reports/tw/4twl
ftp://data.pdbj.org/pub/pdb/validation_reports/tw/4twl Links
Links Assembly
Assembly


 Components
Components Dioscorea japonica (Japanese yam) / Gene: dio5 / Production host:
Dioscorea japonica (Japanese yam) / Gene: dio5 / Production host: 
 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Photon Factory
Photon Factory  / Beamline: BL-5A / Wavelength: 1 Å
/ Beamline: BL-5A / Wavelength: 1 Å Processing
Processing Movie
Movie Controller
Controller












 PDBj
PDBj