[English] 日本語
 Yorodumi
Yorodumi- PDB-4qwc: Ternary Crystal Structures of a Y-family DNA polymerase DPO4 from... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4qwc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ternary Crystal Structures of a Y-family DNA polymerase DPO4 from Sulfobus Solfataricus in Comples with DNA and L-DCDP | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | TRANSFERASE/DNA / Y-FAMILY DNA POLYMERASE / TRANSFERASE-DNA COMPLEX / polymerase | |||||||||
| Function / homology |  Function and homology information Function and homology informationerror-prone translesion synthesis / DNA-templated DNA replication / DNA-directed DNA polymerase / damaged DNA binding / DNA-directed DNA polymerase activity / magnesium ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Saccharolobus solfataricus P2 (archaea) Saccharolobus solfataricus P2 (archaea)Synthetic DNA (others) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.4 Å MOLECULAR REPLACEMENT / Resolution: 2.4 Å | |||||||||
 Authors Authors | Vyas, R. / Suo, Z. | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2014 Journal: Nucleic Acids Res. / Year: 2014Title: Structural and kinetic insights into binding and incorporation of L-nucleotide analogs by a Y-family DNA polymerase. Authors: Gaur, V. / Vyas, R. / Fowler, J.D. / Efthimiopoulos, G. / Feng, J.Y. / Suo, Z. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4qwc.cif.gz 4qwc.cif.gz | 363.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4qwc.ent.gz pdb4qwc.ent.gz | 292.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4qwc.json.gz 4qwc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qw/4qwc https://data.pdbj.org/pub/pdb/validation_reports/qw/4qwc ftp://data.pdbj.org/pub/pdb/validation_reports/qw/4qwc ftp://data.pdbj.org/pub/pdb/validation_reports/qw/4qwc | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4qw8C  4qw9C  4qwaC  4qwbC  4qwdC  4qweC  4fo3 C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AD
| #1: Protein | Mass: 39219.715 Da / Num. of mol.: 2 / Fragment: Dpo4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharolobus solfataricus P2 (archaea) Saccharolobus solfataricus P2 (archaea)Strain: ATCC 35092 / DSM 1617 / JCM 11322 / P2 / Gene: dbh, dpo4, SSO2448 / Plasmid: pET21b / Production host:  |
|---|
-DNA chain , 2 types, 4 molecules BECF
| #2: DNA chain | Mass: 3960.600 Da / Num. of mol.: 2 / Fragment: DNA / Source method: obtained synthetically / Source: (synth.) Synthetic DNA (others) #3: DNA chain | Mass: 5507.565 Da / Num. of mol.: 2 / Fragment: DNA / Source method: obtained synthetically / Source: (synth.) Synthetic DNA (others) |
|---|
-Non-polymers , 4 types, 185 molecules 

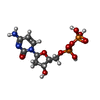




| #4: Chemical | ChemComp-CA / #5: Chemical | #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.72 Å3/Da / Density % sol: 54.75 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 16% PEG 3350, 0.1 M HEPES (PH 7.0), 100MM CALCIUM ACETATE, 2.5% GLYCEROL, VAPOR DIFFUSION, HANGING DROP, TEMPERATURE 298K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 31-ID / Wavelength: 0.97931 / Wavelength: 0.97931 Å / Beamline: 31-ID / Wavelength: 0.97931 / Wavelength: 0.97931 Å |
| Detector | Type: RAYONIX MX225HE / Detector: CCD / Date: Oct 18, 2012 |
| Radiation | Monochromator: KOHZU HLD-4 DOUBLE CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97931 Å / Relative weight: 1 |
| Reflection | Resolution: 2.4→49.22 Å / Num. obs: 41845 / % possible obs: 99.3 % |
| Reflection shell | Resolution: 2.4→2.49 Å / Redundancy: 10 % / Rmerge(I) obs: 0.892 / Mean I/σ(I) obs: 2.8 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4FO3  4fo3 Resolution: 2.4→46.46 Å / Cor.coef. Fo:Fc: 0.946 / Cor.coef. Fo:Fc free: 0.919 / SU B: 23.362 / SU ML: 0.236 / Cross valid method: THROUGHOUT / ESU R: 0.42 / ESU R Free: 0.283 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 70.557 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→46.46 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller

















 PDBj
PDBj








































