Entry Database : PDB / ID : 4o1vTitle SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN Speckle-type POZ protein Keywords / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2 Å Authors Calabrese, M.F. / Watson, E.R. / Schulman, B.A. Journal : Cancer Cell / Year : 2014Title : SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer.Authors: Li, G. / Ci, W. / Karmakar, S. / Chen, K. / Dhar, R. / Fan, Z. / Guo, Z. / Zhang, J. / Ke, Y. / Wang, L. / Zhuang, M. / Hu, S. / Li, X. / Zhou, L. / Li, X. / Calabrese, M.F. / Watson, E.R. / ... Authors : Li, G. / Ci, W. / Karmakar, S. / Chen, K. / Dhar, R. / Fan, Z. / Guo, Z. / Zhang, J. / Ke, Y. / Wang, L. / Zhuang, M. / Hu, S. / Li, X. / Zhou, L. / Li, X. / Calabrese, M.F. / Watson, E.R. / Prasad, S.M. / Rinker-Schaeffer, C. / Eggener, S.E. / Stricker, T. / Tian, Y. / Schulman, B.A. / Liu, J. / White, K.P. History Deposition Dec 16, 2013 Deposition site / Processing site Revision 1.0 Apr 30, 2014 Provider / Type Revision 1.1 Sep 20, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ref_seq_dif Item / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
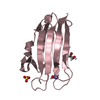
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å
MOLECULAR REPLACEMENT / Resolution: 2 Å  Authors
Authors Citation
Citation Journal: Cancer Cell / Year: 2014
Journal: Cancer Cell / Year: 2014 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4o1v.cif.gz
4o1v.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4o1v.ent.gz
pdb4o1v.ent.gz PDB format
PDB format 4o1v.json.gz
4o1v.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/o1/4o1v
https://data.pdbj.org/pub/pdb/validation_reports/o1/4o1v ftp://data.pdbj.org/pub/pdb/validation_reports/o1/4o1v
ftp://data.pdbj.org/pub/pdb/validation_reports/o1/4o1v
 Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: SPOP / Production host:
Homo sapiens (human) / Gene: SPOP / Production host: 
 Homo sapiens (human) / References: UniProt: P60484
Homo sapiens (human) / References: UniProt: P60484 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 22-BM / Wavelength: 1
/ Beamline: 22-BM / Wavelength: 1  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj















