[English] 日本語
 Yorodumi
Yorodumi- PDB-4np8: Structure of an amyloid forming peptide VQIVYK from the second re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4np8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of an amyloid forming peptide VQIVYK from the second repeat region of tau (alternate polymorph) | |||||||||
 Components Components | Microtubule-associated protein tau | |||||||||
 Keywords Keywords | PROTEIN FIBRIL / amyloid-like protofibril | |||||||||
| Function / homology |  Function and homology information Function and homology informationplus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / generation of neurons ...plus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / generation of neurons / rRNA metabolic process / axonal transport of mitochondrion / regulation of mitochondrial fission / axon development / regulation of microtubule-based movement / regulation of chromosome organization / central nervous system neuron development / intracellular distribution of mitochondria / minor groove of adenine-thymine-rich DNA binding / lipoprotein particle binding / microtubule polymerization / negative regulation of mitochondrial membrane potential / regulation of microtubule polymerization / dynactin binding / apolipoprotein binding / main axon / protein polymerization / axolemma / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / negative regulation of mitochondrial fission / glial cell projection / neurofibrillary tangle assembly / positive regulation of axon extension / regulation of cellular response to heat / Activation of AMPK downstream of NMDARs / positive regulation of superoxide anion generation / positive regulation of protein localization / regulation of long-term synaptic depression / cellular response to brain-derived neurotrophic factor stimulus / positive regulation of microtubule polymerization / supramolecular fiber organization / synapse assembly / cytoplasmic microtubule organization / regulation of calcium-mediated signaling / somatodendritic compartment / axon cytoplasm / phosphatidylinositol binding / astrocyte activation / enzyme inhibitor activity / nuclear periphery / stress granule assembly / protein phosphatase 2A binding / regulation of microtubule cytoskeleton organization / cellular response to reactive oxygen species / Hsp90 protein binding / microglial cell activation / cellular response to nerve growth factor stimulus / synapse organization / PKR-mediated signaling / regulation of synaptic plasticity / protein homooligomerization / regulation of autophagy / response to lead ion / SH3 domain binding / microtubule cytoskeleton organization / memory / cytoplasmic ribonucleoprotein granule / neuron projection development / cell-cell signaling / single-stranded DNA binding / protein-folding chaperone binding / cellular response to heat / microtubule cytoskeleton / actin binding / growth cone / cell body / double-stranded DNA binding / protein-macromolecule adaptor activity / microtubule binding / sequence-specific DNA binding / dendritic spine / amyloid fibril formation / microtubule / learning or memory / neuron projection / membrane raft / negative regulation of gene expression / axon / neuronal cell body / DNA damage response / dendrite / protein kinase binding / enzyme binding / mitochondrion / DNA binding / RNA binding / extracellular region / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.51 Å molecular replacement / Resolution: 1.51 Å | |||||||||
 Authors Authors | Landau, M. / Eisenberg, D. / Sawaya, M.R. / Dannenberg, J. / Kobko, N. | |||||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2009 Journal: Nat.Struct.Mol.Biol. / Year: 2009Title: Molecular mechanisms for protein-encoded inheritance. Authors: Wiltzius, J.J. / Landau, M. / Nelson, R. / Sawaya, M.R. / Apostol, M.I. / Goldschmidt, L. / Soriaga, A.B. / Cascio, D. / Rajashankar, K. / Eisenberg, D. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4np8.cif.gz 4np8.cif.gz | 9.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4np8.ent.gz pdb4np8.ent.gz | 5.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4np8.json.gz 4np8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/np/4np8 https://data.pdbj.org/pub/pdb/validation_reports/np/4np8 ftp://data.pdbj.org/pub/pdb/validation_reports/np/4np8 ftp://data.pdbj.org/pub/pdb/validation_reports/np/4np8 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3fodC  3fpoC  3fr1C  3fthC  3ftkC  3ftlC  3ftrC  3fvaC  2on9S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 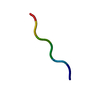
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | x 8
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological unit is a pair of beta sheets. One sheet is constructed from chain A and unit cell translations along the b direction (i.e. X,Y+1,Z; X,Y+2,Z; X,Y+3,Z, etc.). The second sheet is constructed from -1/2-X,1/2+Y,-Z; -1/2-X,3/2+Y,-Z; -1/2-X,5/2+Y,-Z; etc.). |
- Components
Components
| #1: Protein/peptide | Mass: 749.917 Da / Num. of mol.: 1 / Fragment: UNP residues 623-628 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) / References: UniProt: P10636 Homo sapiens (human) / References: UniProt: P10636 |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.56 Å3/Da / Density % sol: 21.29 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: reservoir contained 14% iso-Propanol, 0.07M HEPES-Na pH 7.5, 0.14M Sodium Citrate, and 30% Glycerol, vapor diffusion, hanging drop, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-E / Wavelength: 0.9792 Å / Beamline: 24-ID-E / Wavelength: 0.9792 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Feb 25, 2008 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9792 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→90 Å / Num. all: 825 / Num. obs: 825 / % possible obs: 94.4 % / Observed criterion σ(I): -3 / Redundancy: 4.1 % / Biso Wilson estimate: 15.1 Å2 / Rmerge(I) obs: 0.16 / Net I/σ(I): 8.1 |
| Reflection shell | Resolution: 1.5→1.62 Å / Redundancy: 4 % / Rmerge(I) obs: 0.376 / Mean I/σ(I) obs: 4.7 / Num. unique all: 156 / % possible all: 82.1 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2ON9 Resolution: 1.51→16.78 Å / Cor.coef. Fo:Fc: 0.961 / Cor.coef. Fo:Fc free: 0.955 / WRfactor Rfree: 0.207 / WRfactor Rwork: 0.194 / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.8187 / SU B: 1.895 / SU ML: 0.065 / SU R Cruickshank DPI: 0.0982 / SU Rfree: 0.0894 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.098 / ESU R Free: 0.089 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 26.57 Å2 / Biso mean: 7.5741 Å2 / Biso min: 3.8 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.51→16.78 Å /
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.508→1.547 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller





 PDBj
PDBj





