[English] 日本語
 Yorodumi
Yorodumi- PDB-4m7x: Staphylococcus aureus Type II pantothenate kinase in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4m7x | ||||||
|---|---|---|---|---|---|---|---|
| Title | Staphylococcus aureus Type II pantothenate kinase in complex with a pantothenate analog | ||||||
 Components Components | Type II pantothenate kinase | ||||||
 Keywords Keywords | TRANSFERASE/TRANSFERASE inhibitor / pantothenate kinase / inhibitor / antibacterial / TRANSFERASE-TRANSFERASE inhibitor complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpantothenate kinase / pantothenate kinase activity / coenzyme A biosynthetic process / ATP binding / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.42 Å molecular replacement / Resolution: 1.42 Å | ||||||
 Authors Authors | Mottaghi, K. / Hong, B. / Tempel, W. / Park, H. | ||||||
 Citation Citation |  Journal: ACS Infect Dis / Year: 2016 Journal: ACS Infect Dis / Year: 2016Title: Discovery of Potent Pantothenamide Inhibitors of Staphylococcus aureus Pantothenate Kinase through a Minimal SAR Study: Inhibition Is Due to Trapping of the Product. Authors: Hughes, S.J. / Barnard, L. / Mottaghi, K. / Tempel, W. / Antoshchenko, T. / Hong, B.S. / Allali-Hassani, A. / Smil, D. / Vedadi, M. / Strauss, E. / Park, H.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4m7x.cif.gz 4m7x.cif.gz | 126.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4m7x.ent.gz pdb4m7x.ent.gz | 96.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4m7x.json.gz 4m7x.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/m7/4m7x https://data.pdbj.org/pub/pdb/validation_reports/m7/4m7x ftp://data.pdbj.org/pub/pdb/validation_reports/m7/4m7x ftp://data.pdbj.org/pub/pdb/validation_reports/m7/4m7x | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4m7yC  5elzC  5jicC  2ewsS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Details | AS PER THE AUTHORS THE BIOLOGICAL ASSEMBLY IS UNKNOWN |
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 29955.818 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: MW2 / Gene: coaW, MW2054 / Production host:  |
|---|
-Non-polymers , 5 types, 161 molecules 


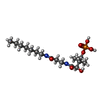





| #2: Chemical | ChemComp-ADP / | ||||
|---|---|---|---|---|---|
| #3: Chemical | ChemComp-MG / | ||||
| #4: Chemical | ChemComp-UNX / #5: Chemical | ChemComp-27Q / | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.4 Å3/Da / Density % sol: 48.4 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion / pH: 8.5 Details: 0.01 M inhibitor, 0.01 M magnesium chloride, 0.01 M ATP were added to the protein sample. 30% PEG4000, 0.2M magnesium chloride, 0.1M Tris hydrochloride, pH 8.5, vapor diffusion, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.97918 Å / Beamline: 19-ID / Wavelength: 0.97918 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Mar 25, 2011 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97918 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.42→68.265 Å / Num. all: 53304 / Num. obs: 53304 / % possible obs: 98.1 % / Redundancy: 9.7 % / Rsym value: 0.086 / Net I/σ(I): 14.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 2EWS Resolution: 1.42→35 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.963 / WRfactor Rfree: 0.205 / WRfactor Rwork: 0.181 / Occupancy max: 1 / Occupancy min: 0.3 / SU B: 1.912 / SU ML: 0.039 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.06 / ESU R Free: 0.061 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: GEOMETRY RESTRAINTS FOR THE INHIBITOR WERE CALCULATED BY PRODRG, AND MODIFIED BASED ON DATA IN THE CAMBRIDGE STRUCTURAL DATABASE. COOT WAS USED FOR MODEL BUILDING. THE MOLPROBITY SERVER WAS ...Details: GEOMETRY RESTRAINTS FOR THE INHIBITOR WERE CALCULATED BY PRODRG, AND MODIFIED BASED ON DATA IN THE CAMBRIDGE STRUCTURAL DATABASE. COOT WAS USED FOR MODEL BUILDING. THE MOLPROBITY SERVER WAS USED FOR MODEL GEOMETRY VALIDATION.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 75.12 Å2 / Biso mean: 23.781 Å2 / Biso min: 11.64 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.42→35 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 17.532 Å / Origin y: 13.8455 Å / Origin z: -3.5562 Å
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



