[English] 日本語
 Yorodumi
Yorodumi- PDB-4ixu: Crystal structure of human Arginase-2 complexed with inhibitor 11... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4ixu | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human Arginase-2 complexed with inhibitor 11d: {(5R)-5-amino-5-carboxy-5-[(3-endo)-8-(3,4-dichlorobenzyl)-8-azabicyclo[3.2.1]oct-3-yl]pentyl}(trihydroxy)borate(1-) | ||||||
 Components Components | Arginase-2, mitochondrial | ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / metalloenzyme / alpha/beta fold / Hydrolase / Arginine metabolism / Manganese / boronate / Mitochondrion / HYDROLASE-HYDROLASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of chemokine (C-C motif) ligand 4 production / negative regulation of activated CD8-positive, alpha-beta T cell apoptotic process / negative regulation of defense response to bacterium / negative regulation of macrophage inflammatory protein 1 alpha production / negative regulation of type 2 immune response / negative regulation of interleukin-13 production / negative regulation of chemokine (C-C motif) ligand 5 production / regulation of interleukin-1 beta production / Urea cycle / arginase ...negative regulation of chemokine (C-C motif) ligand 4 production / negative regulation of activated CD8-positive, alpha-beta T cell apoptotic process / negative regulation of defense response to bacterium / negative regulation of macrophage inflammatory protein 1 alpha production / negative regulation of type 2 immune response / negative regulation of interleukin-13 production / negative regulation of chemokine (C-C motif) ligand 5 production / regulation of interleukin-1 beta production / Urea cycle / arginase / : / arginase activity / urea cycle / negative regulation of CD4-positive, alpha-beta T cell proliferation / negative regulation of interleukin-17 production / ureteric bud development / regulation of reactive oxygen species biosynthetic process / negative regulation of tumor necrosis factor production / striated muscle contraction / nitric oxide biosynthetic process / Mitochondrial protein degradation / positive regulation of cellular senescence / manganese ion binding / adaptive immune response / mitochondrial matrix / innate immune response / mitochondrion / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Cousido-Siah, A. / Mitschler, A. / Ruiz, F.X. / Whitehouse, D. / Beckett, P. / Van Zandt, M.C. / Ji, M.K. / Ryder, T. / Jagdmann, R. / Andreoli, M. ...Cousido-Siah, A. / Mitschler, A. / Ruiz, F.X. / Whitehouse, D. / Beckett, P. / Van Zandt, M.C. / Ji, M.K. / Ryder, T. / Jagdmann, R. / Andreoli, M. / Olczak, J. / Mazur, M. / Czestkowski, W. / Piotrowska, W. / Schroeter, H. / Golebiowski, A. / Podjarny, A. | ||||||
 Citation Citation |  Journal: Bioorg.Med.Chem.Lett. / Year: 2013 Journal: Bioorg.Med.Chem.Lett. / Year: 2013Title: Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction. Authors: Golebiowski, A. / Whitehouse, D. / Beckett, R.P. / Van Zandt, M. / Ji, M.K. / Ryder, T.R. / Jagdmann, E. / Andreoli, M. / Lee, Y. / Sheeler, R. / Conway, B. / Olczak, J. / Mazur, M. / ...Authors: Golebiowski, A. / Whitehouse, D. / Beckett, R.P. / Van Zandt, M. / Ji, M.K. / Ryder, T.R. / Jagdmann, E. / Andreoli, M. / Lee, Y. / Sheeler, R. / Conway, B. / Olczak, J. / Mazur, M. / Czestkowski, W. / Piotrowska, W. / Cousido-Siah, A. / Ruiz, F.X. / Mitschler, A. / Podjarny, A. / Schroeter, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4ixu.cif.gz 4ixu.cif.gz | 212.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4ixu.ent.gz pdb4ixu.ent.gz | 169.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4ixu.json.gz 4ixu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4ixu_validation.pdf.gz 4ixu_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4ixu_full_validation.pdf.gz 4ixu_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  4ixu_validation.xml.gz 4ixu_validation.xml.gz | 44.8 KB | Display | |
| Data in CIF |  4ixu_validation.cif.gz 4ixu_validation.cif.gz | 66.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ix/4ixu https://data.pdbj.org/pub/pdb/validation_reports/ix/4ixu ftp://data.pdbj.org/pub/pdb/validation_reports/ix/4ixu ftp://data.pdbj.org/pub/pdb/validation_reports/ix/4ixu | HTTPS FTP |
-Related structure data
| Related structure data |  4ixvC  1d3vS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
| ||||||||||||
| Details | biological assembly is a homotrimer with one copy in the asymmetric unit |
- Components
Components
-Protein , 1 types, 3 molecules ABC
| #1: Protein | Mass: 33231.656 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ARG2 / Plasmid: pET23b / Production host: Homo sapiens (human) / Gene: ARG2 / Plasmid: pET23b / Production host:  |
|---|
-Non-polymers , 5 types, 816 molecules 


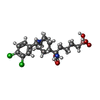





| #2: Chemical | ChemComp-MN / #3: Chemical | #4: Chemical | ChemComp-BME / #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.24 Å3/Da / Density % sol: 62.05 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7 Details: Crystals in condition H3 from Silver Bullet screen (Hampton Research) using Tacsimate in the reservoir, pH 7, VAPOR DIFFUSION, HANGING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 0.9191 Å / Beamline: X06DA / Wavelength: 0.9191 Å |
| Detector | Type: DECTRIS PILATUS 2M / Detector: PIXEL / Date: Jul 8, 2011 |
| Radiation | Monochromator: BARTELS MONOCHROMATOR / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9191 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→46.34 Å / Num. all: 103422 / Num. obs: 103343 / % possible obs: 100 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 6.4 % / Biso Wilson estimate: 24.11 Å2 / Rsym value: 0.082 / Net I/σ(I): 19.3 |
| Reflection shell | Resolution: 1.9→1.97 Å / Redundancy: 6.4 % / Mean I/σ(I) obs: 3.6 / Rsym value: 0.516 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1D3V Resolution: 1.9→46.335 Å / Occupancy max: 1 / Occupancy min: 0 / FOM work R set: 0.8844 / SU ML: 0.15 / Cross valid method: R-free / σ(F): 0 / σ(I): 0 / Phase error: 18.3 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 127.09 Å2 / Biso mean: 19.1735 Å2 / Biso min: 4.53 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→46.335 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 30
|
 Movie
Movie Controller
Controller













 PDBj
PDBj








