[English] 日本語
 Yorodumi
Yorodumi- PDB-4h98: Candida glabrata dihydrofolate reductase complexed with NADPH and... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4h98 | ||||||
|---|---|---|---|---|---|---|---|
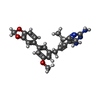
| Title | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-{3-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine (UCP1018) | ||||||
 Components Components | Dihydrofolate Reductase | ||||||
 Keywords Keywords | OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR / Antifungal Agents / Candida glabrata / Drug Design / Enzyme Inhibitors / Fungal Proteins / Tetrahydrofolate Dehydrogenase / OXIDOREDUCTASE-OXIDOREDUCTASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationdihydrofolate metabolic process / dihydrofolate reductase / dihydrofolate reductase activity / folic acid metabolic process / tetrahydrofolate biosynthetic process / one-carbon metabolic process / NADP binding / mitochondrion Similarity search - Function | ||||||
| Biological species |  Candida glabrata (fungus) Candida glabrata (fungus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.9 Å molecular replacement / Resolution: 2.9 Å | ||||||
 Authors Authors | Paulsen, J.L. / Anderson, A.C. | ||||||
 Citation Citation |  Journal: Bioorg.Med.Chem.Lett. / Year: 2013 Journal: Bioorg.Med.Chem.Lett. / Year: 2013Title: Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species. Authors: Paulsen, J.L. / Viswanathan, K. / Wright, D.L. / Anderson, A.C. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR DETERMINED |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4h98.cif.gz 4h98.cif.gz | 107.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4h98.ent.gz pdb4h98.ent.gz | 82 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4h98.json.gz 4h98.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/h9/4h98 https://data.pdbj.org/pub/pdb/validation_reports/h9/4h98 ftp://data.pdbj.org/pub/pdb/validation_reports/h9/4h98 ftp://data.pdbj.org/pub/pdb/validation_reports/h9/4h98 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3ro9C  3roaC  4h95C  4h96C  4h97C  3cseS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||
| 2 | 
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: _ / Ens-ID: 1 / Beg auth comp-ID: VAL / Beg label comp-ID: VAL / End auth comp-ID: HIS / End label comp-ID: HIS / Refine code: _ / Auth seq-ID: 4 - 227 / Label seq-ID: 1 - 224
|
- Components
Components
| #1: Protein | Mass: 26079.883 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Candida glabrata (fungus) Candida glabrata (fungus)Strain: ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65 Gene: CAGL0J03894g, DHFR / Plasmid: pET41 / Production host:  #2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.98 Å3/Da / Density % sol: 38.01 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: PEG 4000, MgCl, Tris, pH 8.5, vapor diffusion, hanging drop, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: May 11, 2011 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.9→42.71 Å / Num. all: 8015 / Num. obs: 8015 / % possible obs: 88.1 % / Observed criterion σ(F): 3 / Observed criterion σ(I): 3 / Redundancy: 2.61 % / Rmerge(I) obs: 0.146 / Net I/σ(I): 5.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3CSE Resolution: 2.9→42.71 Å / Cor.coef. Fo:Fc: 0.913 / Cor.coef. Fo:Fc free: 0.846 / Occupancy max: 1 / Occupancy min: 0.6 / SU B: 22.777 / SU ML: 0.423 / Cross valid method: THROUGHOUT / ESU R Free: 0.622 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.946 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→42.71 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Ens-ID: 1 / Number: 270 / Refine-ID: X-RAY DIFFRACTION / Type: interatomic distance / Rms dev position: 0.09 Å / Weight position: 0.05
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.9→2.976 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller

















 PDBj
PDBj