+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4f2j | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of ZNF217 bound to DNA, P6522 crystal form | ||||||
 Components Components |
| ||||||
 Keywords Keywords | METAL BINDING PROTEIN/DNA / Zinc Finger / Transcription Factor / DNA Binding / Nucleus / METAL BINDING PROTEIN-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationhistone deacetylase complex / Negative Regulation of CDH1 Gene Transcription / DNA-binding transcription repressor activity, RNA polymerase II-specific / Estrogen-dependent gene expression / DNA-binding transcription factor activity, RNA polymerase II-specific / transcription cis-regulatory region binding / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of DNA-templated transcription ...histone deacetylase complex / Negative Regulation of CDH1 Gene Transcription / DNA-binding transcription repressor activity, RNA polymerase II-specific / Estrogen-dependent gene expression / DNA-binding transcription factor activity, RNA polymerase II-specific / transcription cis-regulatory region binding / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of DNA-templated transcription / regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / mitochondrion / zinc ion binding / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SIRAS / Resolution: 2.64 Å SIRAS / Resolution: 2.64 Å | ||||||
 Authors Authors | Vandevenne, M.S. / Jacques, D.A. / Guss, J.M. / Mackay, J.P. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2013 Journal: J.Biol.Chem. / Year: 2013Title: New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217 Authors: Vandevenne, M.S. / Jacques, D.A. / Artuz, C. / Nguyen, C.D. / Kwan, A.H. / Segal, D.J. / Matthews, J.M. / Crossley, M. / Guss, J.M. / Mackay, J.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4f2j.cif.gz 4f2j.cif.gz | 63.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4f2j.ent.gz pdb4f2j.ent.gz | 44.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4f2j.json.gz 4f2j.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/f2/4f2j https://data.pdbj.org/pub/pdb/validation_reports/f2/4f2j ftp://data.pdbj.org/pub/pdb/validation_reports/f2/4f2j ftp://data.pdbj.org/pub/pdb/validation_reports/f2/4f2j | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 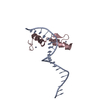
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 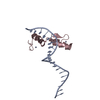
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 6123.977 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: Protein | Mass: 6848.725 Da / Num. of mol.: 1 / Fragment: Zinc fingers 6 and 7 (UNP RESIDUES 469-523) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ZNF217, ZABC1 / Plasmid: pMAL / Production host: Homo sapiens (human) / Gene: ZNF217, ZABC1 / Plasmid: pMAL / Production host:  |
| #3: Chemical |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow |
|
-Data collection
| Diffraction |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Redundancy: 10.4 % / Av σ(I) over netI: 28.64 / Number: 79317 / Rmerge(I) obs: 0.097 / Χ2: 1.06 / D res high: 2.63 Å / D res low: 50 Å / Num. obs: 7637 / % possible obs: 99.5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Diffraction reflection shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.64→99 Å / Num. all: 7517 / Num. obs: 7517 / % possible obs: 98.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 2.5 % / Rmerge(I) obs: 0.091 / Χ2: 3.235 / Net I/σ(I): 9.3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  SIRAS SIRAS | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phasing dm | FOM : 0.57 / FOM acentric: 0.55 / FOM centric: 0.62 / Reflection: 7411 / Reflection acentric: 5383 / Reflection centric: 2028 | ||||||||||||||||||||||||||||||||||||||||||
| Phasing dm shell |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SIRAS / Resolution: 2.64→48.31 Å / Cor.coef. Fo:Fc: 0.924 / Cor.coef. Fo:Fc free: 0.969 / WRfactor Rfree: 0.2778 / WRfactor Rwork: 0.2563 / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.7219 / SU B: 32.121 / SU ML: 0.285 / SU R Cruickshank DPI: 0.3269 / SU Rfree: 0.2467 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.327 / ESU R Free: 0.247 / Stereochemistry target values: MAXIMUM LIKELIHOOD SIRAS / Resolution: 2.64→48.31 Å / Cor.coef. Fo:Fc: 0.924 / Cor.coef. Fo:Fc free: 0.969 / WRfactor Rfree: 0.2778 / WRfactor Rwork: 0.2563 / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.7219 / SU B: 32.121 / SU ML: 0.285 / SU R Cruickshank DPI: 0.3269 / SU Rfree: 0.2467 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.327 / ESU R Free: 0.247 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 68.69 Å2 / Biso mean: 96.517 Å2 / Biso min: 2 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.64→48.31 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.64→2.709 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj









































