+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4d2d | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of a tri peptide bound POT family peptide transporter | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSPORT PROTEIN / MAJOR FACILITATOR SUPERFAMILY / PROTON OLIGOPEPTIDE TRANSPORTER (POT) FAMILY / PEPTIDE TRANSPORTER / TRIPEPTIDE COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationoligopeptide transport / peptide transmembrane transporter activity / identical protein binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  STREPTOCOCCUS THERMOPHILUS (bacteria) STREPTOCOCCUS THERMOPHILUS (bacteria)SYNTHETIC CONSTRUCT (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.522 Å MOLECULAR REPLACEMENT / Resolution: 2.522 Å | ||||||
 Authors Authors | Lyons, J.A. / Parker, J.L. / Solcan, N. / Brinth, A. / Li, D. / Shah, S.T.A. / Caffrey, M. / Newstead, S. | ||||||
 Citation Citation |  Journal: Embo Rep. / Year: 2014 Journal: Embo Rep. / Year: 2014Title: Structural Basis for Polyspecificity in the Pot Family of Proton-Coupled Oligopeptide Transporters. Authors: Lyons, J.A. / Parker, J.L. / Solcan, N. / Brinth, A. / Li, D. / Shah, S.T. / Caffrey, M. / Newstead, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4d2d.cif.gz 4d2d.cif.gz | 106.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4d2d.ent.gz pdb4d2d.ent.gz | 81.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4d2d.json.gz 4d2d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4d2d_validation.pdf.gz 4d2d_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4d2d_full_validation.pdf.gz 4d2d_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  4d2d_validation.xml.gz 4d2d_validation.xml.gz | 19.4 KB | Display | |
| Data in CIF |  4d2d_validation.cif.gz 4d2d_validation.cif.gz | 26.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d2/4d2d https://data.pdbj.org/pub/pdb/validation_reports/d2/4d2d ftp://data.pdbj.org/pub/pdb/validation_reports/d2/4d2d ftp://data.pdbj.org/pub/pdb/validation_reports/d2/4d2d | HTTPS FTP |
-Related structure data
| Related structure data |  4d2bC  4d2cC  4apsS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Protein/peptide , 2 types, 2 molecules AB
| #1: Protein | Mass: 53721.125 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  STREPTOCOCCUS THERMOPHILUS (bacteria) / Strain: LMG 18311 / Plasmid: PWALDO-GFPD / Production host: STREPTOCOCCUS THERMOPHILUS (bacteria) / Strain: LMG 18311 / Plasmid: PWALDO-GFPD / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 231.249 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) SYNTHETIC CONSTRUCT (others) |
-Non-polymers , 4 types, 52 molecules 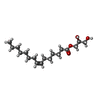
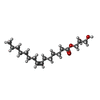





| #3: Chemical | | #4: Chemical | #5: Chemical | ChemComp-PO4 / | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.95 Å3/Da / Density % sol: 58.3 % / Description: NONE |
|---|---|
| Crystal grow | Method: lipidic cubic phase / pH: 7 Details: 16-23 %(V/V) PEG 400, 0.1 M HEPES PH 7.0, 0.15-0.55 M NH4H2PO4 AND 10 MM TRI-ALANINE. CRYSTALS WERE GROWN BY THE LCP METHOD WITH 7.8 MAG AS HOSTING LIPID. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 23-ID-B / Wavelength: 1.0332 / Beamline: 23-ID-B / Wavelength: 1.0332 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Dec 2, 2012 / Details: K-B PAIR OF BIOMORPH MIRRORS |
| Radiation | Monochromator: DOUBLE CRYSTAL MONOCHROMATOR SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0332 Å / Relative weight: 1 |
| Reflection | Resolution: 2.52→75.56 Å / Num. obs: 21341 / % possible obs: 98.7 % / Observed criterion σ(I): -3 / Redundancy: 3.2 % / Biso Wilson estimate: 44.26 Å2 / Rmerge(I) obs: 0.08 / Net I/σ(I): 10.4 |
| Reflection shell | Resolution: 2.52→2.59 Å / Redundancy: 3.2 % / Rmerge(I) obs: 0.57 / Mean I/σ(I) obs: 2.2 / % possible all: 99.3 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 4APS Resolution: 2.522→75.556 Å / SU ML: 0.29 / σ(F): 1.34 / Phase error: 24.67 / Stereochemistry target values: ML Details: RESIDUES 1-5, 272-275, 414-419, 479-491 ARE DISORDERED TRIPEPTIDE WAS MODELED WITH ITS C-TERMINUS ORIENTATED TOWARDS THE EXTRACELLULAR SIDE. SEE PAPER FOR FURTHER DISCUSSION.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 50.229 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.522→75.556 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj





