[English] 日本語
 Yorodumi
Yorodumi- PDB-6yof: Structure of PepTSt from COC IMISX setup collected by rotation se... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6yof | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of PepTSt from COC IMISX setup collected by rotation serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | ||||||
 Components Components | Di-or tripeptide:H+ symporter | ||||||
 Keywords Keywords | TRANSPORT PROTEIN / 2D X-ray phase-contrast imaging / IMISX / in situ rotation images / prelocation / still images / serial crystallography / HYDROLASE | ||||||
| Function / homology |  Function and homology information Function and homology informationtripeptide transmembrane transport / tripeptide transmembrane transporter activity / peptide:proton symporter activity / dipeptide transmembrane transporter activity / identical protein binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  Streptococcus thermophilus (bacteria) Streptococcus thermophilus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.45 Å MOLECULAR REPLACEMENT / Resolution: 2.45 Å | ||||||
 Authors Authors | Huang, C.-Y. / Martiel, I. / Villanueva-Perez, P. / Panepucci, E. / Caffrey, M. / Wang, M. | ||||||
 Citation Citation |  Journal: Iucrj / Year: 2020 Journal: Iucrj / Year: 2020Title: Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography. Authors: Martiel, I. / Huang, C.Y. / Villanueva-Perez, P. / Panepucci, E. / Basu, S. / Caffrey, M. / Pedrini, B. / Bunk, O. / Stampanoni, M. / Wang, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6yof.cif.gz 6yof.cif.gz | 116.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6yof.ent.gz pdb6yof.ent.gz | 90.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6yof.json.gz 6yof.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yo/6yof https://data.pdbj.org/pub/pdb/validation_reports/yo/6yof ftp://data.pdbj.org/pub/pdb/validation_reports/yo/6yof ftp://data.pdbj.org/pub/pdb/validation_reports/yo/6yof | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6yobC  6yocC  6yodC  6yoeC  6yogC  5d58S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 52782.148 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) (bacteria) Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) (bacteria)Gene: dtpT, stu0970 / Production host:  |
|---|
-Non-polymers , 5 types, 96 molecules 
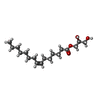







| #2: Chemical | ChemComp-PO4 / | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-78M / ( #4: Chemical | ChemComp-PG0 / | #5: Chemical | ChemComp-PGE / | #6: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.14 Å3/Da / Density % sol: 60.79 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase / pH: 7 Details: 250-325 mM NH4H2PO4, 100 mM HEPES, pH 7.0, 21-22 %(v/v) PEG 400 and 10 mM Ala-Phe |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: Y |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 1 Å / Beamline: X06SA / Wavelength: 1 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Dec 4, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.45→49.55 Å / Num. obs: 23572 / % possible obs: 99.95 % / Redundancy: 52.17 % / CC1/2: 0.99 / Rrim(I) all: 0.47 / Net I/σ(I): 13.01 |
| Reflection shell | Resolution: 2.45→2.51 Å / Redundancy: 46.37 % / Mean I/σ(I) obs: 1.34 / Num. unique obs: 1819 / CC1/2: 0.56 / Rrim(I) all: 70.4 / % possible all: 100 |
| Serial crystallography sample delivery | Method: fixed target |
| Serial crystallography sample delivery fixed target | Description: IMISX / Sample dehydration prevention: COC sandwich / Sample holding: IMISX / Support base: 3D-printed holder in standard goniometer base |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5D58 Resolution: 2.45→49.55 Å / SU ML: 0.36 / Cross valid method: THROUGHOUT / σ(F): 1.35 / Phase error: 26.94
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 154.83 Å2 / Biso mean: 69.8842 Å2 / Biso min: 33.44 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.45→49.55 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 8 / % reflection obs: 100 %
|
 Movie
Movie Controller
Controller












 PDBj
PDBj






