[English] 日本語
 Yorodumi
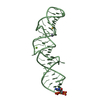
Yorodumi- PDB-4by2: SAS-4 (dCPAP) TCP domain in complex with a Proline Rich Motif of ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4by2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | SAS-4 (dCPAP) TCP domain in complex with a Proline Rich Motif of Ana2 (dSTIL) of Drosophila Melanogaster | ||||||
 Components Components | ANASTRAL SPINDLE 2, SAS 4 | ||||||
 Keywords Keywords | STRUCTURAL PROTEIN / CPAP / STIL / MICROCEPHALY / MCPH / CENTRIOLE / TCP DOMAIN / EXTENDED BETA SHEET | ||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to pericentriolar material / ciliary basal body organization / centriole elongation / syncytial blastoderm mitotic cell cycle / centriole assembly / asymmetric cell division / centrosome separation / centriole-centriole cohesion / microtubule anchoring at centrosome / asymmetric neuroblast division ...protein localization to pericentriolar material / ciliary basal body organization / centriole elongation / syncytial blastoderm mitotic cell cycle / centriole assembly / asymmetric cell division / centrosome separation / centriole-centriole cohesion / microtubule anchoring at centrosome / asymmetric neuroblast division / male meiotic nuclear division / locomotion / centrosome cycle / pericentriolar material / centriole replication / microtubule polymerization / establishment of mitotic spindle orientation / cilium assembly / centriole / tubulin binding / molecular adaptor activity / ciliary basal body / centrosome / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.57 Å MAD / Resolution: 2.57 Å | ||||||
 Authors Authors | Cottee, M.A. / Muschalik, N. / Wong, Y.L. / Johnson, C.M. / Johnson, S. / Andreeva, A. / Oegema, K. / Lea, S.M. / Raff, J.W. / van Breugel, M. | ||||||
 Citation Citation |  Journal: Elife / Year: 2013 Journal: Elife / Year: 2013Title: Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Authors: Cottee, M.A. / Muschalik, N. / Wong, Y.L. / Johnson, C.M. / Johnson, S. / Andreeva, A. / Oegema, K. / Lea, S.M. / Raff, J.W. / van Breugel, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4by2.cif.gz 4by2.cif.gz | 109.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4by2.ent.gz pdb4by2.ent.gz | 83.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4by2.json.gz 4by2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/by/4by2 https://data.pdbj.org/pub/pdb/validation_reports/by/4by2 ftp://data.pdbj.org/pub/pdb/validation_reports/by/4by2 ftp://data.pdbj.org/pub/pdb/validation_reports/by/4by2 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Protein | Mass: 28606.064 Da / Num. of mol.: 3 Fragment: PROLINE-RICH-MOTIF RESIDUES 2-47 BOUND TO TCP DOMAIN, RESIDUES 700-901 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Description: CODON-OPTIMISED SEQUENCE BASED ON UNIPROT ENTRIES Q9XZ31 (ANA2) AND Q9VI72 (SAS-4) Production host:  #2: Chemical | ChemComp-EDO / | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.09 Å3/Da / Density % sol: 60.25 % / Description: NONE |
|---|---|
| Crystal grow | Method: vapor diffusion, sitting drop / pH: 6.5 Details: 100 MM MES/IMIDAZOLE MIX PH 6.5, 30 MM MGCL2, 30 MM CACL2, 20 % ETHYLENE GLYCOL, 10 % PEG LIQUOR, IN A 0.2UL SITTING DROP. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9795 / Beamline: I04 / Wavelength: 0.9795 |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Sep 23, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 2.57→64.6 Å / Num. obs: 31911 / % possible obs: 97.6 % / Observed criterion σ(I): 2 / Redundancy: 3.1 % / Biso Wilson estimate: 71.41 Å2 / Rmerge(I) obs: 0.09 / Net I/σ(I): 7.6 |
| Reflection shell | Resolution: 2.57→8.9 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.51 / Mean I/σ(I) obs: 1.7 / % possible all: 97.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD MADStarting model: NONE Resolution: 2.57→64.6 Å / Cor.coef. Fo:Fc: 0.8539 / Cor.coef. Fo:Fc free: 0.836 / SU R Cruickshank DPI: 0.307 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.287 / SU Rfree Blow DPI: 0.226 / SU Rfree Cruickshank DPI: 0.237
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 70.8 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.474 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.57→64.6 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.57→2.65 Å / Total num. of bins used: 16
|
 Movie
Movie Controller
Controller














 PDBj
PDBj


