[English] 日本語
 Yorodumi
Yorodumi- PDB-3wkn: Crystal structure of the artificial protein AFFinger p17 (AF.p17)... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3wkn | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
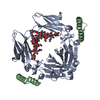
| Title | Crystal structure of the artificial protein AFFinger p17 (AF.p17) complexed with Fc fragment of human IgG | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | IMMUNE SYSTEM / immunoglobulin-like beta-sandwich / binding protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationFc-gamma receptor I complex binding / complement-dependent cytotoxicity / antibody-dependent cellular cytotoxicity / immunoglobulin receptor binding / IgG immunoglobulin complex / immunoglobulin complex, circulating / Classical antibody-mediated complement activation / Initial triggering of complement / FCGR activation / complement activation, classical pathway ...Fc-gamma receptor I complex binding / complement-dependent cytotoxicity / antibody-dependent cellular cytotoxicity / immunoglobulin receptor binding / IgG immunoglobulin complex / immunoglobulin complex, circulating / Classical antibody-mediated complement activation / Initial triggering of complement / FCGR activation / complement activation, classical pathway / Role of phospholipids in phagocytosis / antigen binding / FCGR3A-mediated IL10 synthesis / Regulation of Complement cascade / B cell receptor signaling pathway / FCGR3A-mediated phagocytosis / Regulation of actin dynamics for phagocytic cup formation / antibacterial humoral response / Interleukin-4 and Interleukin-13 signaling / blood microparticle / adaptive immune response / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å | |||||||||
 Authors Authors | Watanabe, H. / Honda, S. | |||||||||
 Citation Citation |  Journal: Chem.Biol. / Year: 2015 Journal: Chem.Biol. / Year: 2015Title: Adaptive Assembly: Maximizing the Potential of a Given Functional Peptide with a Tailor-Made Protein Scaffold. Authors: Watanabe, H. / Honda, S. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3wkn.cif.gz 3wkn.cif.gz | 421.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3wkn.ent.gz pdb3wkn.ent.gz | 351.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3wkn.json.gz 3wkn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wk/3wkn https://data.pdbj.org/pub/pdb/validation_reports/wk/3wkn ftp://data.pdbj.org/pub/pdb/validation_reports/wk/3wkn ftp://data.pdbj.org/pub/pdb/validation_reports/wk/3wkn | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 23975.072 Da / Num. of mol.: 8 / Fragment: UNP residues 119-330 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: IGHG1 / Cell (production host): ovary cell / Production host: Homo sapiens (human) / Gene: IGHG1 / Cell (production host): ovary cell / Production host:  #2: Protein | Mass: 6122.594 Da / Num. of mol.: 8 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host:  #3: Polysaccharide | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose- ...beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.82 Å3/Da / Density % sol: 56.37 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 5.6 Details: 40% PEG 4000, 0.1M sodium citrate, 0.2M ammonium acetate, pH 5.6, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: AR-NW12A / Wavelength: 1 Å / Beamline: AR-NW12A / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Oct 12, 2011 |
| Radiation | Monochromator: double Si(111) monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→50 Å / Num. all: 68126 / Num. obs: 68016 / Observed criterion σ(I): -3 / Redundancy: 7.4 % / Rmerge(I) obs: 0.066 |
| Reflection shell | Resolution: 2.8→2.85 Å / Redundancy: 7 % / Rmerge(I) obs: 0.389 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.9→20 Å / Cor.coef. Fo:Fc: 0.93 / Cor.coef. Fo:Fc free: 0.901 / SU B: 14.323 / SU ML: 0.273 / Cross valid method: THROUGHOUT / ESU R Free: 0.389 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS MOLECULAR REPLACEMENT / Resolution: 2.9→20 Å / Cor.coef. Fo:Fc: 0.93 / Cor.coef. Fo:Fc free: 0.901 / SU B: 14.323 / SU ML: 0.273 / Cross valid method: THROUGHOUT / ESU R Free: 0.389 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 50.215 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller









 PDBj
PDBj







