[English] 日本語
 Yorodumi
Yorodumi- PDB-3szx: Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Rev... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3szx | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations | ||||||||||||||||||
 Components Components | RNA (5'-R(P* Keywords KeywordsRNA / CUG Repeat RNA | Function / homology | RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.204 Å MOLECULAR REPLACEMENT / Resolution: 2.204 Å  Authors AuthorsKumar, A. / Park, H. / Pengfei, F. / Parkesh, R. / Guo, M. / Nettles, K.W. / Disney, M.D. |  Citation Citation Journal: Biochemistry / Year: 2011 Journal: Biochemistry / Year: 2011Title: Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations Authors: Kumar, A. / Park, H. / Fang, P. / Parkesh, R. / Guo, M. / Nettles, K.W. / Disney, M.D. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3szx.cif.gz 3szx.cif.gz | 41.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3szx.ent.gz pdb3szx.ent.gz | 29.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3szx.json.gz 3szx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sz/3szx https://data.pdbj.org/pub/pdb/validation_reports/sz/3szx ftp://data.pdbj.org/pub/pdb/validation_reports/sz/3szx ftp://data.pdbj.org/pub/pdb/validation_reports/sz/3szx | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 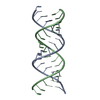
| |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: RNA chain | Mass: 6039.569 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: Polyribonucleotide #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.94 Å3/Da / Density % sol: 36.53 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 6 Details: 10 mM magnesium sulfate, 50 mM sodium cacodylate, 1.8 M lithium sulfate , pH 6.0, VAPOR DIFFUSION, SITTING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 0.97855 Å / Beamline: BL9-2 / Wavelength: 0.97855 Å | ||||||||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Jan 9, 2011 / Details: Rh coated flat mirror, toroidal focusing mirror | ||||||||||||||||||||||||||||||
| Radiation | Monochromator: Double crystal monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97855 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.2→33.41 Å / Num. all: 4571 / Num. obs: 4571 / % possible obs: 99.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 | ||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.204→33.41 Å / SU ML: 0.32 / σ(F): 0.13 / Phase error: 35.2 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 2.204→33.41 Å / SU ML: 0.32 / σ(F): 0.13 / Phase error: 35.2 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.05 Å / VDW probe radii: 0.3 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 42.097 Å2 / ksol: 0.462 e/Å3 | ||||||||||||||||||||||||||||
| Displacement parameters |
| ||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.204→33.41 Å
| ||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj






























