[English] 日本語
 Yorodumi
Yorodumi- PDB-3stk: Crystal Structure of human LFABP complex with two molecules of pa... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3stk | ||||||
|---|---|---|---|---|---|---|---|
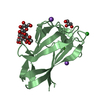
| Title | Crystal Structure of human LFABP complex with two molecules of palmitic acid (holo-LFABP) | ||||||
 Components Components | Fatty acid-binding protein, liver | ||||||
 Keywords Keywords | LIPID BINDING PROTEIN / LFABP / Palmitic acid / Fatty acid binding | ||||||
| Function / homology |  Function and homology information Function and homology informationcellular detoxification / Heme degradation / Triglyceride catabolism / antioxidant activity / peroxisomal matrix / fatty acid transport / Regulation of lipid metabolism by PPARalpha / fatty acid binding / PPARA activates gene expression / Cytoprotection by HMOX1 ...cellular detoxification / Heme degradation / Triglyceride catabolism / antioxidant activity / peroxisomal matrix / fatty acid transport / Regulation of lipid metabolism by PPARalpha / fatty acid binding / PPARA activates gene expression / Cytoprotection by HMOX1 / cellular response to hydrogen peroxide / cellular response to hypoxia / chromatin binding / extracellular exosome / nucleoplasm / nucleus / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.55 Å molecular replacement / Resolution: 1.55 Å | ||||||
 Authors Authors | Sharma, A. / Sharma, A. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Fatty acid induced remodeling within the Human liver fatty acid binding protein Authors: Sharma, A. / Sharma, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3stk.cif.gz 3stk.cif.gz | 44.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3stk.ent.gz pdb3stk.ent.gz | 30.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3stk.json.gz 3stk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/st/3stk https://data.pdbj.org/pub/pdb/validation_reports/st/3stk ftp://data.pdbj.org/pub/pdb/validation_reports/st/3stk ftp://data.pdbj.org/pub/pdb/validation_reports/st/3stk | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14599.728 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FABP1, FABPL / Production host: Homo sapiens (human) / Gene: FABP1, FABPL / Production host:  | ||
|---|---|---|---|
| #2: Chemical | | #3: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.22 Å3/Da / Density % sol: 44.51 % / Mosaicity: 1.716 ° |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 8 Details: 0.15M potassium bromide, 30% polyethylene glycol monomethyl ether (PEG MME) 2000, pH 8.0, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.54 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.54 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MAR scanner 345 mm plate / Detector: IMAGE PLATE / Date: May 5, 2010 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.54→50 Å / Num. obs: 18383 / % possible obs: 92.2 % / Redundancy: 10.7 % / Rmerge(I) obs: 0.046 / Χ2: 1.978 / Net I/σ(I): 24 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.55→25.193 Å / Occupancy max: 1 / Occupancy min: 0.89 / FOM work R set: 0.8738 / SU ML: 0.41 / σ(F): 1.33 / Phase error: 19.1 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.55→25.193 Å / Occupancy max: 1 / Occupancy min: 0.89 / FOM work R set: 0.8738 / SU ML: 0.41 / σ(F): 1.33 / Phase error: 19.1 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.72 Å / VDW probe radii: 1 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 45.979 Å2 / ksol: 0.341 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 54.52 Å2 / Biso mean: 25.135 Å2 / Biso min: 13.17 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.55→25.193 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 7
|
 Movie
Movie Controller
Controller












 PDBj
PDBj













