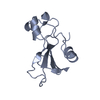
Entry Database : PDB / ID : 3q9vTitle Crystal structure of rra c-terminal domain(123-221) from Deinococcus radiodurans DNA-binding response regulator Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Deinococcus radiodurans (radioresistant)Method / / / Resolution : 1.60174363532 Å Authors Liu, Y. / Gao, Z.Q. / Dong, Y.H. / Ji, C.N. Funding support Organization Grant number Country National Basic Research Program of China (973 Program) 2009CB918600
Journal : To be Published Title : Crystal structure of rra c-terminal domain (123-221) from Deinococcus radiodurans.Authors : Liu, Y. / Gao, Z.Q. / Dong, Y.H. / Ji, C.N. History Deposition Jan 10, 2011 Deposition site / Processing site Revision 1.0 Jan 11, 2012 Provider / Type Revision 1.1 Nov 8, 2017 Group / Category / Item Revision 1.2 Feb 23, 2022 Group / Derived calculationsCategory database_2 / pdbx_struct_assembly ... database_2 / pdbx_struct_assembly / pdbx_struct_assembly_gen / pdbx_struct_oper_list / struct_ref_seq_dif Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_assembly.details / _pdbx_struct_assembly.method_details / _pdbx_struct_assembly.oligomeric_count / _pdbx_struct_assembly.oligomeric_details / _struct_ref_seq_dif.details Revision 1.3 Jul 27, 2022 Group Advisory / Author supporting evidence ... Advisory / Author supporting evidence / Data collection / Database references / Other / Refinement description / Source and taxonomy / Structure summary Category diffrn / entity ... diffrn / entity / entity_src_gen / pdbx_audit_support / pdbx_contact_author / pdbx_database_status / pdbx_distant_solvent_atoms / refine / refine_ls_shell / reflns / reflns_shell / struct / struct_ref_seq_dif Item _diffrn.pdbx_serial_crystal_experiment / _entity.details ... _diffrn.pdbx_serial_crystal_experiment / _entity.details / _entity_src_gen.gene_src_strain / _entity_src_gen.pdbx_beg_seq_num / _entity_src_gen.pdbx_end_seq_num / _entity_src_gen.pdbx_host_org_ncbi_taxonomy_id / _entity_src_gen.pdbx_host_org_scientific_name / _entity_src_gen.pdbx_host_org_strain / _entity_src_gen.pdbx_seq_type / _pdbx_contact_author.id / _pdbx_contact_author.identifier_ORCID / _pdbx_contact_author.name_salutation / _pdbx_database_status.SG_entry / _refine.occupancy_max / _refine.occupancy_min / _refine.overall_FOM_work_R_set / _refine.pdbx_ls_cross_valid_method / _refine_ls_shell.d_res_high / _refine_ls_shell.d_res_low / _refine_ls_shell.number_reflns_all / _refine_ls_shell.pdbx_total_number_of_bins_used / _reflns.number_all / _reflns.pdbx_netI_over_sigmaI / _reflns_shell.number_unique_all / _reflns_shell.number_unique_obs / _struct.pdbx_CASP_flag / _struct_ref_seq_dif.details Revision 2.0 Aug 3, 2022 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Derived calculations / Experimental preparation / Other / Refinement description / Structure summary Category atom_site / atom_site_anisotrop ... atom_site / atom_site_anisotrop / atom_sites / cell / entity / exptl_crystal_grow / pdbx_distant_solvent_atoms / pdbx_nonpoly_scheme / pdbx_refine_tls / pdbx_refine_tls_group / pdbx_struct_assembly / pdbx_struct_assembly_gen / pdbx_struct_assembly_prop / pdbx_struct_sheet_hbond / pdbx_struct_special_symmetry / pdbx_unobs_or_zero_occ_atoms / pdbx_validate_symm_contact / pdbx_validate_torsion / refine / refine_hist / refine_ls_restr / refine_ls_shell / reflns / software / struct_conf / struct_mon_prot_cis / struct_sheet / struct_sheet_order / struct_sheet_range / symmetry Item _atom_sites.fract_transf_matrix[1][3] / _cell.volume ... _atom_sites.fract_transf_matrix[1][3] / _cell.volume / _entity.pdbx_number_of_molecules / _exptl_crystal_grow.pdbx_details / _refine.B_iso_max / _refine.B_iso_mean / _refine.B_iso_min / _refine.aniso_B[1][1] / _refine.aniso_B[1][2] / _refine.aniso_B[1][3] / _refine.aniso_B[2][2] / _refine.aniso_B[2][3] / _refine.aniso_B[3][3] / _refine.ls_R_factor_R_free / _refine.ls_R_factor_R_work / _refine.ls_R_factor_all / _refine.ls_R_factor_obs / _refine.ls_d_res_high / _refine.ls_d_res_low / _refine.ls_number_reflns_R_work / _refine.ls_number_reflns_all / _refine.ls_percent_reflns_R_free / _refine.ls_percent_reflns_obs / _refine.overall_SU_ML / _refine.pdbx_ls_sigma_F / _refine.pdbx_overall_phase_error / _refine.pdbx_stereochemistry_target_values / _refine.solvent_model_param_bsol / _refine.solvent_model_param_ksol / _refine_hist.d_res_high / _refine_hist.d_res_low / _refine_hist.number_atoms_solvent / _refine_hist.number_atoms_total / _refine_ls_restr.dev_ideal / _refine_ls_shell.R_factor_R_free / _refine_ls_shell.R_factor_R_work / _refine_ls_shell.d_res_high / _refine_ls_shell.d_res_low / _refine_ls_shell.number_reflns_R_work / _refine_ls_shell.percent_reflns_obs / _reflns.B_iso_Wilson_estimate / _struct_conf.beg_auth_comp_id / _struct_conf.beg_auth_seq_id / _struct_conf.beg_label_comp_id / _struct_conf.beg_label_seq_id / _struct_conf.end_auth_comp_id / _struct_conf.end_auth_seq_id / _struct_conf.end_label_comp_id / _struct_conf.end_label_seq_id / _struct_conf.pdbx_PDB_helix_id / _struct_conf.pdbx_PDB_helix_length / _symmetry.space_group_name_Hall Revision 2.1 Aug 17, 2022 Group / Category / Item Revision 2.2 Nov 1, 2023 Group / Refinement descriptionCategory / chem_comp_bond / pdbx_initial_refinement_model
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Deinococcus radiodurans (radioresistant)
Deinococcus radiodurans (radioresistant) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.60174363532 Å
MOLECULAR REPLACEMENT / Resolution: 1.60174363532 Å  Authors
Authors China, 1items
China, 1items  Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3q9v.cif.gz
3q9v.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3q9v.ent.gz
pdb3q9v.ent.gz PDB format
PDB format 3q9v.json.gz
3q9v.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/q9/3q9v
https://data.pdbj.org/pub/pdb/validation_reports/q9/3q9v ftp://data.pdbj.org/pub/pdb/validation_reports/q9/3q9v
ftp://data.pdbj.org/pub/pdb/validation_reports/q9/3q9v
 Links
Links Assembly
Assembly

 Components
Components Deinococcus radiodurans (radioresistant)
Deinococcus radiodurans (radioresistant)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site: BSRF
SYNCHROTRON / Site: BSRF  / Beamline: 3W1A / Wavelength: 1 Å
/ Beamline: 3W1A / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj


