[English] 日本語
 Yorodumi
Yorodumi- PDB-3q75: Cryptococcus neoformans protein farnesyltransferase in complex wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3q75 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryptococcus neoformans protein farnesyltransferase in complex with FPT-II and TKCVVM peptide | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE / Protein prenyltransferase | ||||||
| Function / homology |  Function and homology information Function and homology informationprotein prenyltransferase activity / protein farnesyltransferase / protein farnesyltransferase activity / protein farnesyltransferase complex / zinc ion binding Similarity search - Function | ||||||
| Biological species |  Cryptococcus neoformans (Cryptococcus neoformans serotype A) Cryptococcus neoformans (Cryptococcus neoformans serotype A) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.14 Å MOLECULAR REPLACEMENT / Resolution: 2.14 Å | ||||||
 Authors Authors | Hast, M.A. / Beese, L.S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Structures of Cryptococcus neoformans Protein Farnesyltransferase Reveal Strategies for Developing Inhibitors That Target Fungal Pathogens. Authors: Hast, M.A. / Nichols, C.B. / Armstrong, S.M. / Kelly, S.M. / Hellinga, H.W. / Alspaugh, J.A. / Beese, L.S. | ||||||
| History |
| ||||||
| Remark 999 | There is no UniPort database reference sequence available at the time of the deposition. The ...There is no UniPort database reference sequence available at the time of the deposition. The sequences presented here are of farnesyltransferase alpha subunit(residues 15-349) and beta subunit from Cryptococcus neoformans var. grubii strain H99. The first 14 residues in the alpha subunit(chain A) represent expression tag. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3q75.cif.gz 3q75.cif.gz | 331.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3q75.ent.gz pdb3q75.ent.gz | 267 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3q75.json.gz 3q75.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q7/3q75 https://data.pdbj.org/pub/pdb/validation_reports/q7/3q75 ftp://data.pdbj.org/pub/pdb/validation_reports/q7/3q75 ftp://data.pdbj.org/pub/pdb/validation_reports/q7/3q75 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3q73C  3q78C  3q79C  3q7aC  3q7fC  3sfxC  3sfyC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
-Farnesyltransferase ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 40913.234 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Cryptococcus neoformans (Cryptococcus neoformans serotype A) Cryptococcus neoformans (Cryptococcus neoformans serotype A)Strain: H99 / Production host:  |
|---|---|
| #2: Protein | Mass: 56806.879 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Cryptococcus neoformans (Cryptococcus neoformans serotype A) Cryptococcus neoformans (Cryptococcus neoformans serotype A)Strain: H99 / Production host:  |
-Protein/peptide , 1 types, 1 molecules G
| #3: Protein/peptide | Mass: 680.899 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: synthetic peptide substrate |
|---|
-Non-polymers , 4 types, 478 molecules 
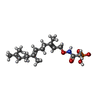





| #4: Chemical | ChemComp-ZN / | ||
|---|---|---|---|
| #5: Chemical | ChemComp-FII / [( | ||
| #6: Chemical | | #7: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.33 Å3/Da / Density % sol: 63.08 % |
|---|---|
| Crystal grow | Temperature: 290 K / pH: 9.5 Details: 15% PEG4K, 100mM CAPSO pH 9.5, 100mM NaCl, 150mM Lithium sulfate, VAPOR DIFFUSION, HANGING DROP, temperature 290K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 12.3.1 / Wavelength: 1.116 / Beamline: 12.3.1 / Wavelength: 1.116 |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Feb 10, 2010 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.116 Å / Relative weight: 1 |
| Reflection | Resolution: 2.14→50 Å / Num. obs: 70375 / % possible obs: 95.8 % / Observed criterion σ(I): -3 |
| Reflection shell | Resolution: 2.14→2.19 Å / % possible all: 96.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.14→45.45 Å / Cor.coef. Fo:Fc: 0.941 / Cor.coef. Fo:Fc free: 0.923 / SU B: 9.299 / SU ML: 0.113 / Cross valid method: THROUGHOUT / ESU R Free: 0.169 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS MOLECULAR REPLACEMENT / Resolution: 2.14→45.45 Å / Cor.coef. Fo:Fc: 0.941 / Cor.coef. Fo:Fc free: 0.923 / SU B: 9.299 / SU ML: 0.113 / Cross valid method: THROUGHOUT / ESU R Free: 0.169 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.92 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.14→45.45 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.14→2.193 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




